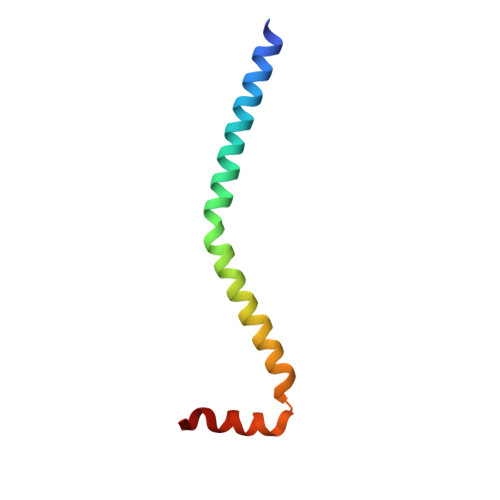
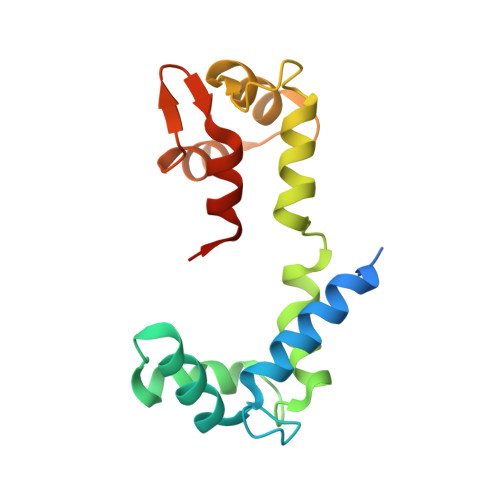
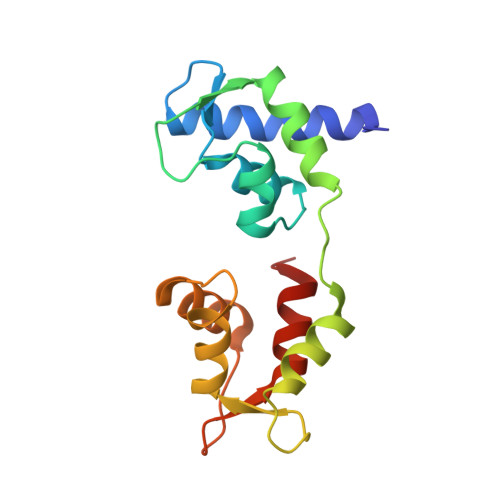
Crystal structure of a phosphorylated light chain domain of scallop smooth-muscle Myosin.
Senthil Kumar, V.S., O'Neall-Hennessey, E., Reshetnikova, L., Brown, J.H., Robinson, H., Szent-Gyorgyi, A.G., Cohen, C.(2011) Biophys J 101: 2185-2189
- PubMed: 22067157
- DOI: https://doi.org/10.1016/j.bpj.2011.09.028
- Primary Citation of Related Structures:
3TS5, 3TUY - PubMed Abstract:
We have determined the crystal structure of a phosphorylated smooth-muscle myosin light chain domain (LCD). This reconstituted LCD is of a sea scallop catch muscle myosin with its phosphorylatable regulatory light chain (RLC SmoA). In the crystal structure, Arg(16), an arginine residue that is present in this isoform but not in vertebrate smooth-muscle RLC, stabilizes the phosphorylation site. This arginine interacts with the carbonyl group of the phosphorylation-site serine in the unphosphorylated LCD (determined previously), and with the phosphate group when the serine is phosphorylated. However, the overall conformation of the LCD is essentially unchanged upon phosphorylation. This result provides additional evidence that phosphorylation of the RLC is unlikely to act as an on-switch in regulation of scallop catch muscle myosin.
- Rosenstiel Basic Medical Sciences Research Center, Brandeis University, Waltham, Massachusetts, USA.
Organizational Affiliation: