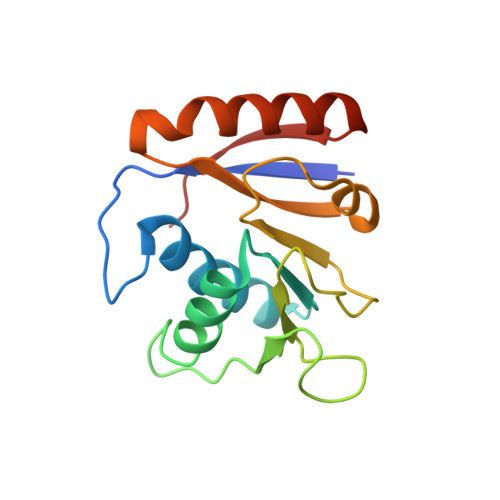
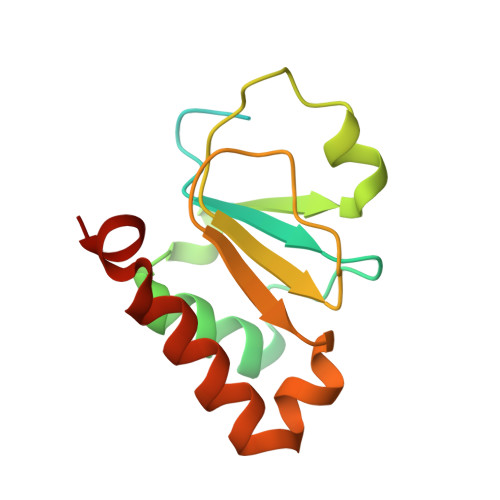
Structure of the SSB-DNA polymerase III interface and its role in DNA replication.
Marceau, A.H., Bahng, S., Massoni, S.C., George, N.P., Sandler, S.J., Marians, K.J., Keck, J.L.(2011) EMBO J 30: 4236-4247
- PubMed: 21857649
- DOI: https://doi.org/10.1038/emboj.2011.305
- Primary Citation of Related Structures:
3SXU - PubMed Abstract:
Interactions between single-stranded DNA-binding proteins (SSBs) and the DNA replication machinery are found in all organisms, but the roles of these contacts remain poorly defined. In Escherichia coli, SSB's association with the χ subunit of the DNA polymerase III holoenzyme has been proposed to confer stability to the replisome and to aid delivery of primers to the lagging-strand DNA polymerase. Here, the SSB-binding site on χ is identified crystallographically and biochemical and cellular studies delineate the consequences of destabilizing the χ/SSB interface. An essential role for the χ/SSB interaction in lagging-strand primer utilization is not supported. However, sequence changes in χ that block complex formation with SSB lead to salt-dependent uncoupling of leading- and lagging-strand DNA synthesis and to a surprising obstruction of the leading-strand DNA polymerase in vitro, pointing to roles for the χ/SSB complex in replisome establishment and maintenance. Destabilization of the χ/SSB complex in vivo produces cells with temperature-dependent cell cycle defects that appear to arise from replisome instability.
- Department of Biomolecular Chemistry, University of Wisconsin School of Medicine and Public Health, Madison, WI 53706-1532, USA.
Organizational Affiliation: