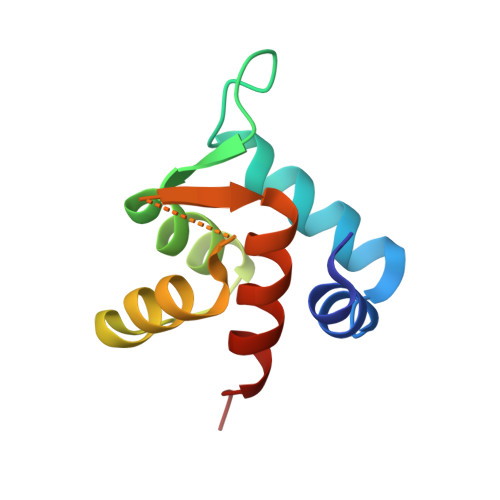
The structure of cardiac troponin C regulatory domain with bound Cd(2+) reveals a closed conformation and unique ion coordination.
Zhang, X.L., Tibbits, G.F., Paetzel, M.(2013) Acta Crystallogr D Biol Crystallogr 69: 722-734
- PubMed: 23633581
- DOI: https://doi.org/10.1107/S0907444913001182
- Primary Citation of Related Structures:
3SD6, 3SWB, 4GJE, 4GJF, 4GJG - PubMed Abstract:
The amino-terminal domain of cardiac troponin C (cNTnC) is an essential Ca(2+) sensor found in cardiomyocytes. It undergoes a conformational change upon Ca(2+) binding and transduces the signal to the rest of the troponin complex to initiate cardiac muscle contraction. Two classical EF-hand motifs (EF1 and EF2) are present in cNTnC. Under physiological conditions, only EF2 binds Ca(2+); EF1 is a vestigial site that has lost its function in binding Ca(2+) owing to amino-acid sequence changes during evolution. Proteins with EF-hand motifs are capable of binding divalent cations other than calcium. Here, the crystal structure of wild-type (WT) human cNTnC in complex with Cd(2+) is presented. The structure of Cd(2+)-bound cNTnC with the disease-related mutation L29Q, as well as a structure with the residue differences D2N, V28I, L29Q and G30D (NIQD), which have been shown to have functional importance in Ca(2+) sensing at lower temperatures in ectothermic species, have also been determined. The structures resemble the overall conformation of NMR structures of Ca(2+)-bound cNTnC, but differ significantly from a previous crystal structure of Cd(2+)-bound cNTnC in complex with deoxycholic acid. The subtle structural changes observed in the region near the mutations may play a role in the increased Ca(2+) affinity. The 1.4 Å resolution WT cNTnC structure, which is the highest resolution structure yet obtained for cardiac troponin C, reveals a Cd(2+) ion coordinated in the canonical pentagonal bipyramidal geometry in EF2 despite three residues in the loop being disordered. A Cd(2+) ion found in the vestigial ion-binding site of EF1 is coordinated in a noncanonical `distorted' octahedral geometry. A comparison of the ion coordination observed within EF-hand-containing proteins for which structures have been solved in the presence of Cd(2+) is presented. A refolded WT cNTnC structure is also presented.
- Department of Molecular Biology and Biochemistry, Simon Fraser University, South Science Building, 8888 University Drive, Burnaby, British Columbia V5A 1S6, Canada.
Organizational Affiliation: