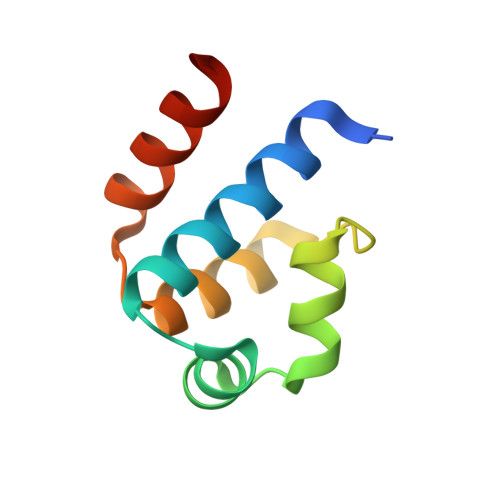
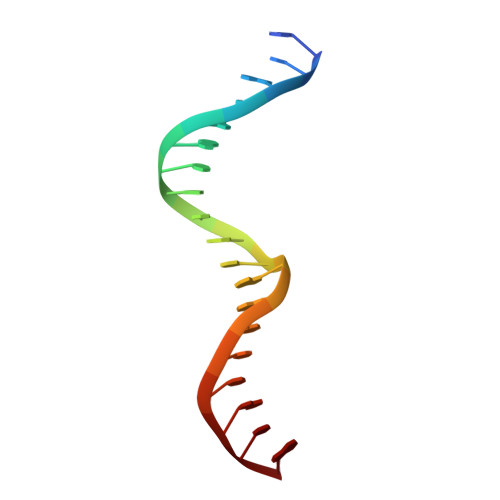
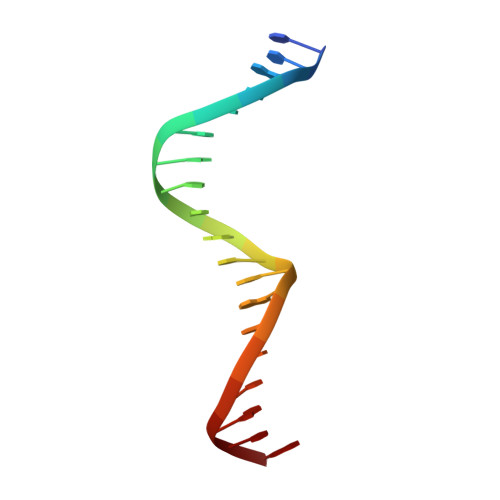
Recognition of dual symmetry by the controller protein C.Esp1396I based on the structure of the transcriptional activation complex.
McGeehan, J.E., Ball, N.J., Streeter, S.D., Thresh, S.J., Kneale, G.G.(2012) Nucleic Acids Res 40: 4158-4167
- PubMed: 22210861
- DOI: https://doi.org/10.1093/nar/gkr1250
- Primary Citation of Related Structures:
3S8Q - PubMed Abstract:
The controller protein C.Esp1396I regulates the timing of gene expression of the restriction-modification (RM) genes of the RM system Esp1396I. The molecular recognition of promoter sequences by such transcriptional regulators is poorly understood, in part because the DNA sequence motifs do not conform to a well-defined symmetry. We report here the crystal structure of the controller protein bound to a DNA operator site. The structure reveals how two different symmetries within the operator are simultaneously recognized by the homo-dimeric protein, underpinned by a conformational change in one of the protein subunits. The recognition of two different DNA symmetries through movement of a flexible loop in one of the protein subunits may represent a general mechanism for the recognition of pseudo-symmetric DNA sequences.
- Biomolecular Structure Group, Institute of Biomedical and Biomolecular Sciences, School of Biological Sciences, University of Portsmouth, Portsmouth, Hampshire PO1 2DY, UK.
Organizational Affiliation: