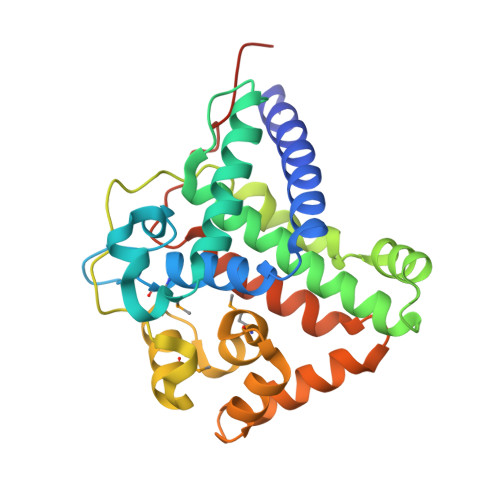
Crystal structure of the TL29 protein from Arabidopsis thaliana: An APX homolog without peroxidase activity.
Lundberg, E., Storm, P., Schroder, W.P., Funk, C.(2011) J Struct Biol 176: 24-31
- PubMed: 21798352
- DOI: https://doi.org/10.1016/j.jsb.2011.07.004
- Primary Citation of Related Structures:
3RRW - PubMed Abstract:
TL29 is a plant-specific protein found in the thylakoid lumen of chloroplasts. Despite the putative requirement in plants for a peroxidase close to the site of photosynthetic oxygen production, and the sequence homology of TL29 to ascorbate peroxidases, so far biochemical methods have not shown this enzyme to possess peroxidase activity. Here we report the three-dimensional X-ray crystal structure of recombinant TL29 from Arabidopsis thaliana at a resolution of 2.5Å. The overall structure of TL29 is mainly alpha helical with six longer and six shorter helical segments. The TL29 structure resembles that of typical ascorbate peroxidases, however, crucial differences were found in regions that would be important for heme and ascorbate binding. Such differences suggest it to be highly unlikely that TL29 functions as a peroxidase.
- Department of Chemistry, Umeå University, SE-90187 Umeå, Sweden.
Organizational Affiliation: