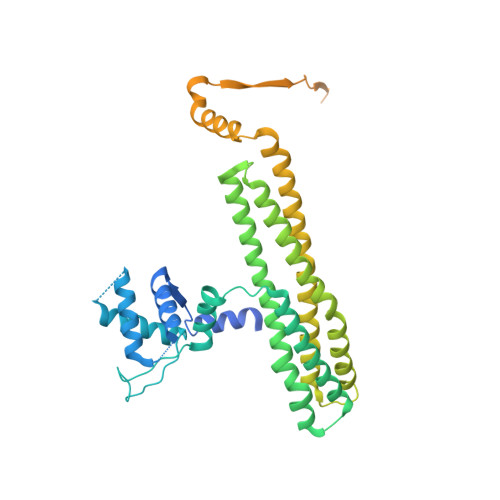
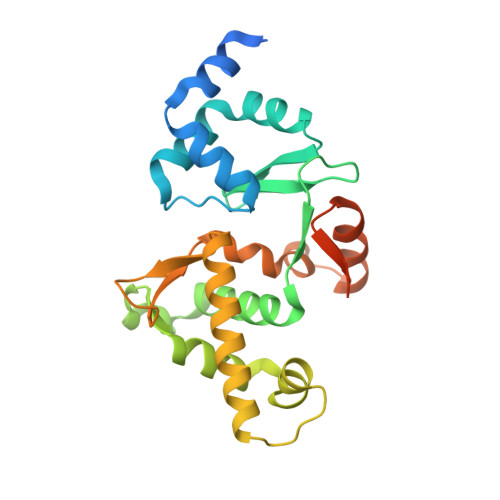
The role of MukE in assembling a functional MukBEF complex.
Gloyd, M., Ghirlando, R., Guarne, A.(2011) J Mol Biology 412: 578-590
- PubMed: 21855551
- DOI: https://doi.org/10.1016/j.jmb.2011.08.009
- Primary Citation of Related Structures:
3RPU - PubMed Abstract:
The MukB-MukE-MukF protein complex is essential for chromosome condensation and segregation in Escherichia coli. The central component of this complex, the MukB protein, is related functionally and structurally to the ubiquitous SMC (structural maintenance of chromosomes) proteins. In a manner similar to SMC, MukB requires the association of two accessory proteins (MukE and MukF) for its function. MukF is a constitutive dimer that bridges the interaction between MukB and MukE. While MukB can condense DNA on its own, it requires MukF and MukE to ensure proper chromosome segregation. Here, we present a novel structure of the E. coli MukE-MukF complex, in which the intricate crystal packing interactions reveal an alternative MukE dimerization interface spanning both N- and C-terminal winged-helix domains of the protein. The structure also unveils additional cross-linking interactions between adjacent MukE-MukF complexes mediated by MukE. A variant of MukE encompassing point mutations on one of these surfaces does not affect assembly of the MukB-MukE-MukF complex and yet cannot restore the temperature sensitivity of the mukE∷kan strain, suggesting that this surface may mediate critical protein-protein interactions between MukB-MukE-MukF complexes. Since the dimerization interface of MukE overlaps with the region of the protein that interacts with MukB in the MukB-MukE-MukF complex, we suggest that competing MukB-MukE and MukE-MukE interactions may regulate the formation of higher-order structures of bacterial condensin.
- Department of Biochemistry and Biomedical Sciences, HSC-4N57A, McMaster University, Hamilton, ON, Canada L8S 4K1.
Organizational Affiliation: