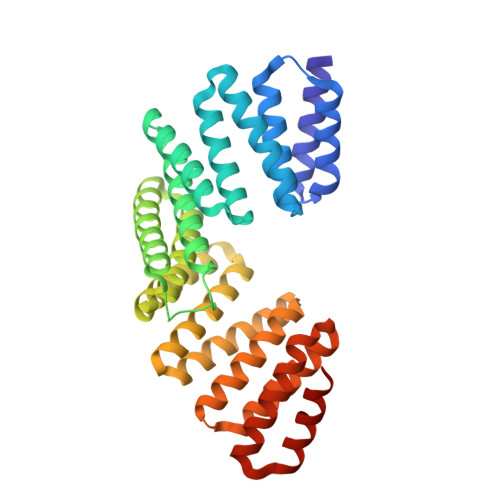
LGN/mInsc and LGN/NuMA complex structures suggest distinct functions in asymmetric cell division for the Par3/mInsc/LGN and G[alpha]i/LGN/NuMA pathways
Zhu, J., Wen, W., Zheng, Z., Shang, Y., Wei, Z., Xiao, Z., Pan, Z., Du, Q., Wang, W., Zhang, M.(2011) Mol Cell 43: 418-431
- PubMed: 21816348
- DOI: https://doi.org/10.1016/j.molcel.2011.07.011
- Primary Citation of Related Structures:
3RO2, 3RO3 - PubMed Abstract:
Asymmetric cell division requires the establishment of cortical cell polarity and the orientation of the mitotic spindle along the axis of cell polarity. Evidence from invertebrates demonstrates that the Par3/Par6/aPKC and NuMA/LGN/Gαi complexes, which are thought to be physically linked by the adaptor protein mInscuteable (mInsc), play indispensable roles in this process. However, the molecular basis for the binding of LGN to NuMA and mInsc is poorly understood. The high-resolution structures of the LGN/NuMA and LGN/mInsc complexes presented here provide mechanistic insights into the distinct and highly specific interactions of the LGN TPRs with mInsc and NuMA. Structural comparisons, together with biochemical and cell biology studies, demonstrate that the interactions of NuMA and mInsc with LGN are mutually exclusive, with mInsc binding preferentially. Our results suggest that the Par3/mInsc/LGN and NuMA/LGN/Gαi complexes play sequential and partially overlapping roles in asymmetric cell division.
- Department of Chemistry, Shanghai Key Laboratory of Molecular Catalysis and Innovative Materials, Fudan University, Shanghai, PR China.
Organizational Affiliation: