Mutagenesis of tryptophan199 reveals that electron hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis
Jensen, L.M.R., Wilmot, C.M.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylamine utilization protein MauG | 373 | Paracoccus denitrificans PD1222 | Mutation(s): 1 Gene Names: mauG, Pden_4736 EC: 1 | 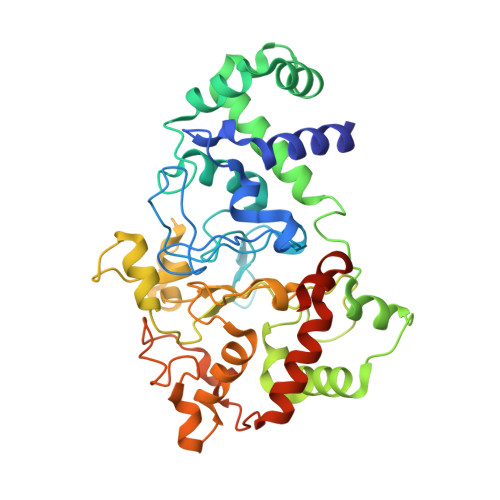 | |
UniProt | |||||
Find proteins for Q51658 (Paracoccus denitrificans (strain Pd 1222)) Explore Q51658 Go to UniProtKB: Q51658 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q51658 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylamine dehydrogenase light chain | 137 | Paracoccus denitrificans PD1222 | Mutation(s): 0 Gene Names: Pden_4733 EC: 1.4.99.3 (PDB Primary Data), 1.4.9.1 (UniProt) | 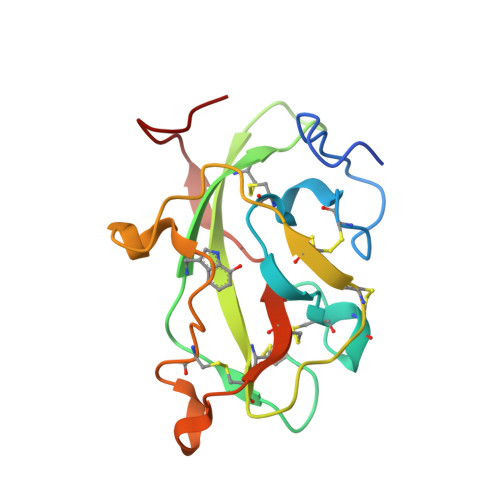 | |
UniProt | |||||
Find proteins for A1BBA0 (Paracoccus denitrificans (strain Pd 1222)) Explore A1BBA0 Go to UniProtKB: A1BBA0 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1BBA0 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Methylamine dehydrogenase heavy chain | 386 | Paracoccus denitrificans PD1222 | Mutation(s): 0 Gene Names: Pden_4730 EC: 1.4.99.3 (PDB Primary Data), 1.4.9.1 (UniProt) | 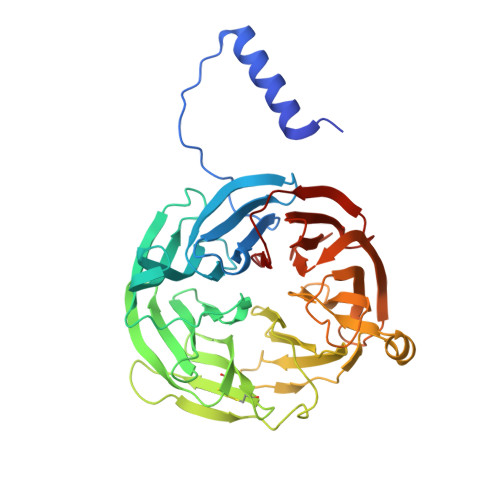 | |
UniProt | |||||
Find proteins for A1BB97 (Paracoccus denitrificans (strain Pd 1222)) Explore A1BB97 Go to UniProtKB: A1BB97 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | A1BB97 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 7 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| HEC Query on HEC | J [auth A], K [auth A], O [auth B], P [auth B] | HEME C C34 H34 Fe N4 O4 HXQIYSLZKNYNMH-LJNAALQVSA-N |  | ||
| 1PE Query on 1PE | R [auth F] | PENTAETHYLENE GLYCOL C10 H22 O6 JLFNLZLINWHATN-UHFFFAOYSA-N |  | ||
| PG4 Query on PG4 | S [auth F] | TETRAETHYLENE GLYCOL C8 H18 O5 UWHCKJMYHZGTIT-UHFFFAOYSA-N |  | ||
| EDO Query on EDO | Q [auth B] | 1,2-ETHANEDIOL C2 H6 O2 LYCAIKOWRPUZTN-UHFFFAOYSA-N |  | ||
| ACT Query on ACT | T [auth F] | ACETATE ION C2 H3 O2 QTBSBXVTEAMEQO-UHFFFAOYSA-M |  | ||
| CA Query on CA | G [auth A], L [auth B] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| NA Query on NA | H [auth A], I [auth A], M [auth B], N [auth B] | SODIUM ION Na FKNQFGJONOIPTF-UHFFFAOYSA-N |  | ||
| Modified Residues 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Type | Formula | 2D Diagram | Parent |
| 0AF Query on 0AF | C, E | L-PEPTIDE LINKING | C11 H12 N2 O3 |  | TRP |
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.527 | α = 109.94 |
| b = 83.524 | β = 91.54 |
| c = 107.782 | γ = 105.78 |
| Software Name | Purpose |
|---|---|
| Blu-Ice | data collection |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| REFMAC | phasing |