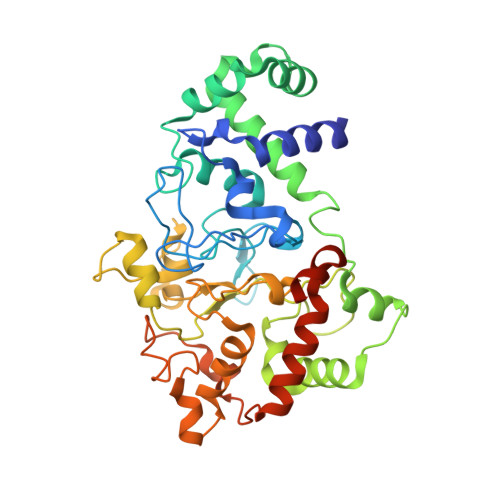
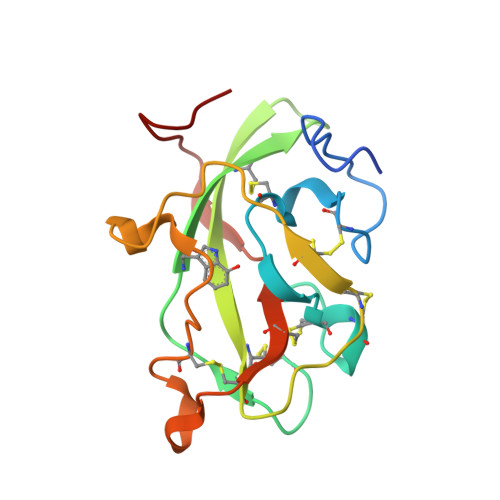
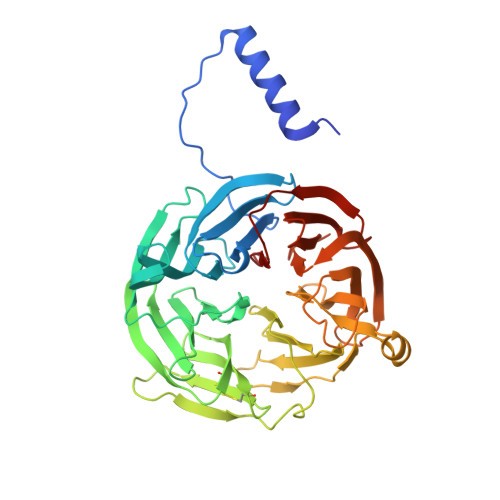
Mutagenesis of tryptophan199 suggests that hopping is required for MauG-dependent tryptophan tryptophylquinone biosynthesis.
Tarboush, N.A., Jensen, L.M., Yukl, E.T., Geng, J., Liu, A., Wilmot, C.M., Davidson, V.L.(2011) Proc Natl Acad Sci U S A 108: 16956-16961
- PubMed: 21969534
- DOI: https://doi.org/10.1073/pnas.1109423108
- Primary Citation of Related Structures:
3RLM, 3RMZ, 3RN0 - PubMed Abstract:
The diheme enzyme MauG catalyzes the posttranslational modification of the precursor protein of methylamine dehydrogenase (preMADH) to complete biosynthesis of its protein-derived tryptophan tryptophylquinone (TTQ) cofactor. Catalysis proceeds through a high valent bis-Fe(IV) redox state and requires long-range electron transfer (ET), as the distance between the modified residues of preMADH and the nearest heme iron of MauG is 19.4 Å. Trp199 of MauG resides at the MauG-preMADH interface, positioned midway between the residues that are modified and the nearest heme. W199F and W199K mutations did not affect the spectroscopic and redox properties of MauG, or its ability to stabilize the bis-Fe(IV) state. Crystal structures of complexes of W199F/K MauG with preMADH showed no significant perturbation of the MauG-preMADH structure or protein interface. However, neither MauG variant was able to synthesize TTQ from preMADH. In contrast, an ET reaction from diferrous MauG to quinone MADH, which does not require the bis-Fe(IV) intermediate, was minimally affected by the W199F/K mutations. W199F/K MauGs were able to oxidize quinol MADH to form TTQ, the putative final two-electron oxidation of the biosynthetic process, but with k(cat)/K(m) values approximately 10% that of wild-type MauG. The differential effects of the W199F/K mutations on these three different reactions are explained by a critical role for Trp199 in mediating multistep hopping from preMADH to bis-Fe(IV) MauG during the long-range ET that is required for TTQ biosynthesis.
- Department of Biochemistry, University of Mississippi Medical Center, Jackson, MS 39216, USA.
Organizational Affiliation: