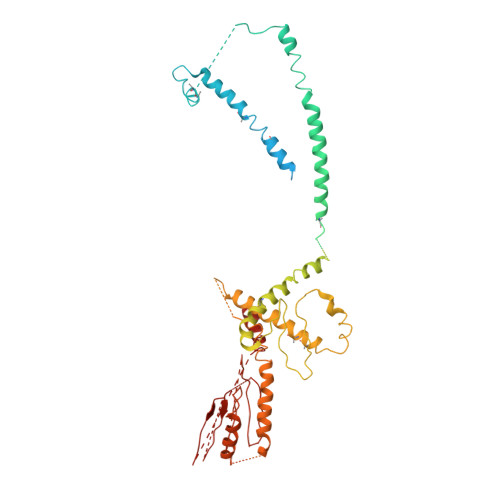
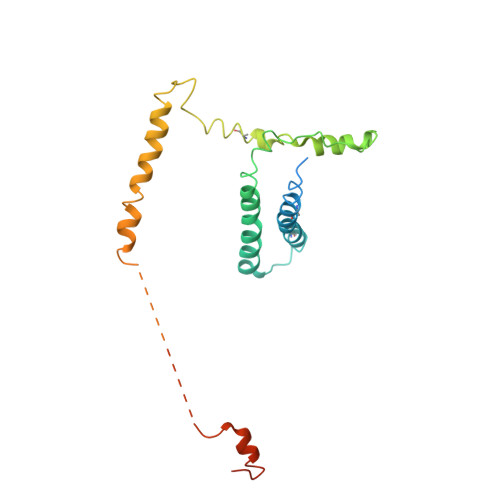
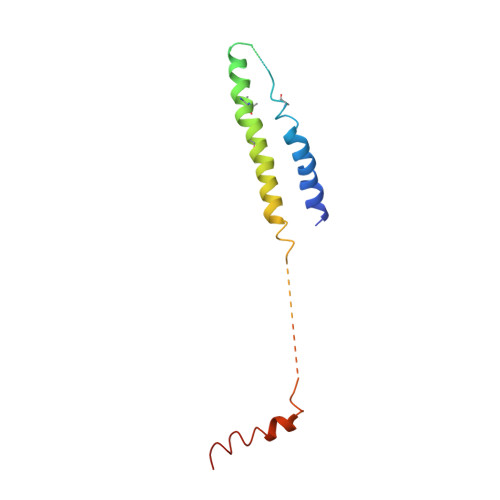
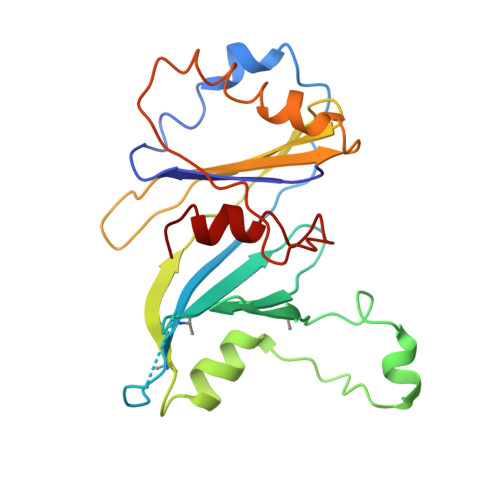
Architecture of the Mediator head module.
Imasaki, T., Calero, G., Cai, G., Tsai, K.L., Yamada, K., Cardelli, F., Erdjument-Bromage, H., Tempst, P., Berger, I., Kornberg, G.L., Asturias, F.J., Kornberg, R.D., Takagi, Y.(2011) Nature 475: 240-243
- PubMed: 21725323
- DOI: https://doi.org/10.1038/nature10162
- Primary Citation of Related Structures:
3RJ1 - PubMed Abstract:
Mediator is a key regulator of eukaryotic transcription, connecting activators and repressors bound to regulatory DNA elements with RNA polymerase II (Pol II). In the yeast Saccharomyces cerevisiae, Mediator comprises 25 subunits with a total mass of more than one megadalton (refs 5, 6) and is organized into three modules, called head, middle/arm and tail. Our understanding of Mediator assembly and its role in regulating transcription has been impeded so far by limited structural information. Here we report the crystal structure of the essential Mediator head module (seven subunits, with a mass of 223 kilodaltons) at a resolution of 4.3 ångströms. Our structure reveals three distinct domains, with the integrity of the complex centred on a bundle of ten helices from five different head subunits. An intricate pattern of interactions within this helical bundle ensures the stable assembly of the head subunits and provides the binding sites for general transcription factors and Pol II. Our structural and functional data suggest that the head module juxtaposes transcription factor IIH and the carboxy-terminal domain of the largest subunit of Pol II, thereby facilitating phosphorylation of the carboxy-terminal domain of Pol II. Our results reveal architectural principles underlying the role of Mediator in the regulation of gene expression.
- Department of Biochemistry and Molecular Biology, Indiana University School of Medicine, 635 Barnhill Drive, Indianapolis, Indiana 46202, USA.
Organizational Affiliation: