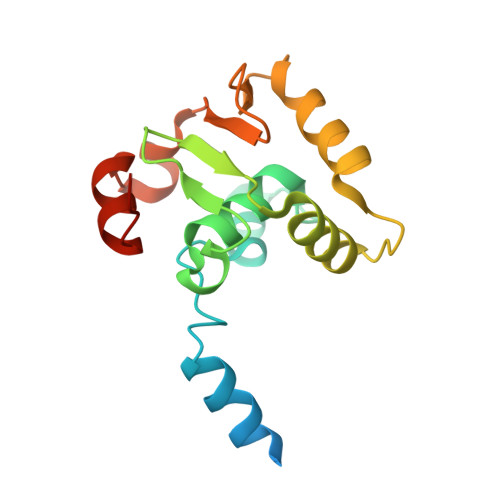
Structural basis of ets1 cooperative binding to widely separated sites on promoter DNA.
Babayeva, N.D., Baranovskaya, O.I., Tahirov, T.H.(2012) PLoS One 7: e33698-e33698
- PubMed: 22432043
- DOI: https://doi.org/10.1371/journal.pone.0033698
- Primary Citation of Related Structures:
3RI4 - PubMed Abstract:
Ets1 is a member of the Ets family of transcription factors. Ets1 is expressed in autoinhibited form and its DNA binding depends on partner proteins bound to adjacent sequences or the relative positioning of a second Ets-binding site (EBS). The autoinhibition of Ets1 is mediated by structural coupling of regions flanking the DNA-binding domain. The NMR structure of Ets1 revealed that the inhibitory regions comprised of helices HI1 and HI2 and H4 are packed together on the Ets domain to form an inhibitory module. The crystal structure of Ets1 unexpectedly revealed a homodimer in which homodimerisation occurs via swapping of HI1 helices. Modeling of DNA binding indicates that the Ets1 dimer can bind to two antiparallel pieces of DNA. To verify this, we crystallized and solved the structure of the complex comprised of Ets1 dimer and two pieces of DNA. DNA binding by Ets1 dimer resulted in formation of additional intermolecular protein•DNA interactions, implying that the complex formation is cooperative.
- Eppley Institute for Research in Cancer and Allied Diseases, University of Nebraska Medical Center, Omaha, Nebraska, United States of America.
Organizational Affiliation: