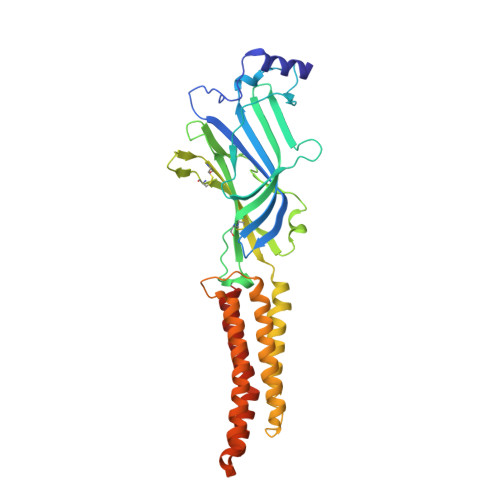
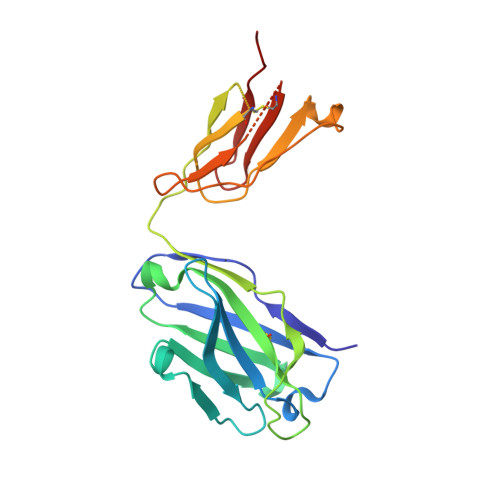
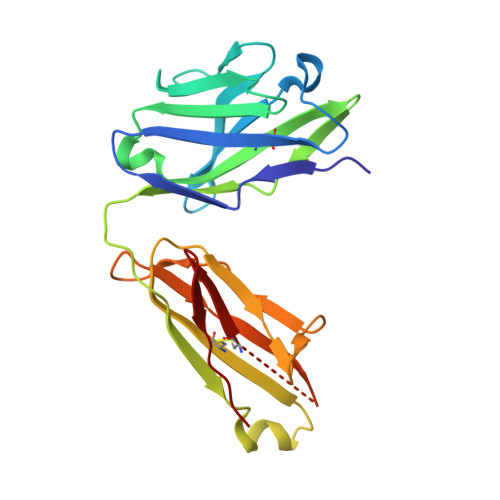
Principles of activation and permeation in an anion-selective Cys-loop receptor.
Hibbs, R.E., Gouaux, E.(2011) Nature 474: 54-60
- PubMed: 21572436
- DOI: https://doi.org/10.1038/nature10139
- Primary Citation of Related Structures:
3RHW, 3RI5, 3RIA, 3RIF - PubMed Abstract:
Fast inhibitory neurotransmission is essential for nervous system function and is mediated by binding of inhibitory neurotransmitters to receptors of the Cys-loop family embedded in the membranes of neurons. Neurotransmitter binding triggers a conformational change in the receptor, opening an intrinsic chloride channel and thereby dampening neuronal excitability. Here we present the first three-dimensional structure, to our knowledge, of an inhibitory anion-selective Cys-loop receptor, the homopentameric Caenorhabditis elegans glutamate-gated chloride channel α (GluCl), at 3.3 Å resolution. The X-ray structure of the GluCl-Fab complex was determined with the allosteric agonist ivermectin and in additional structures with the endogenous neurotransmitter L-glutamate and the open-channel blocker picrotoxin. Ivermectin, used to treat river blindness, binds in the transmembrane domain of the receptor and stabilizes an open-pore conformation. Glutamate binds in the classical agonist site at subunit interfaces, and picrotoxin directly occludes the pore near its cytosolic base. GluCl provides a framework for understanding mechanisms of fast inhibitory neurotransmission and allosteric modulation of Cys-loop receptors.
- Vollum Institute, Oregon Health and Science University, 3181 SW Sam Jackson Park Road, Portland, Oregon 97239, USA.
Organizational Affiliation: