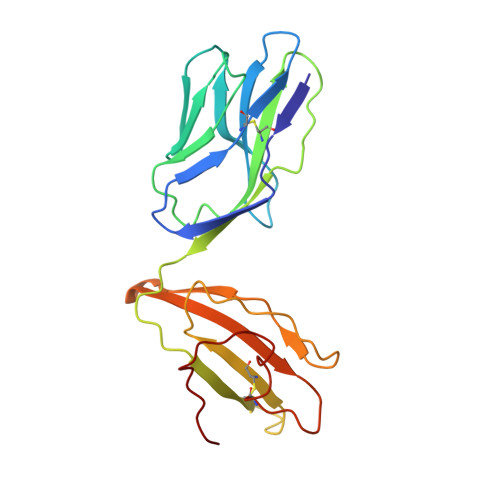
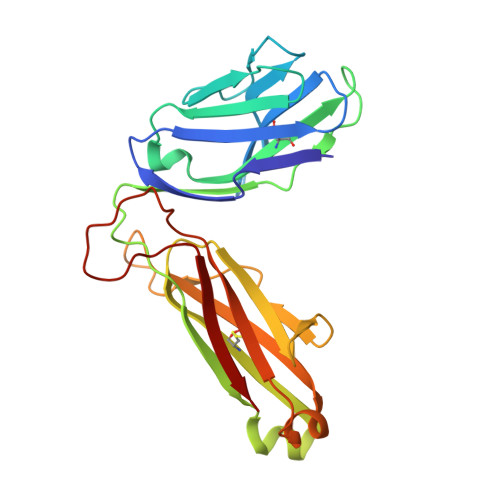
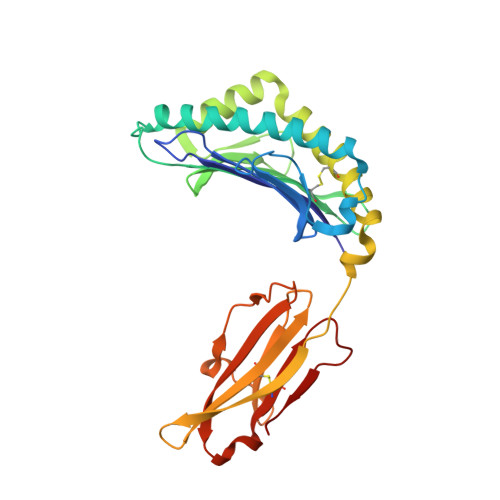
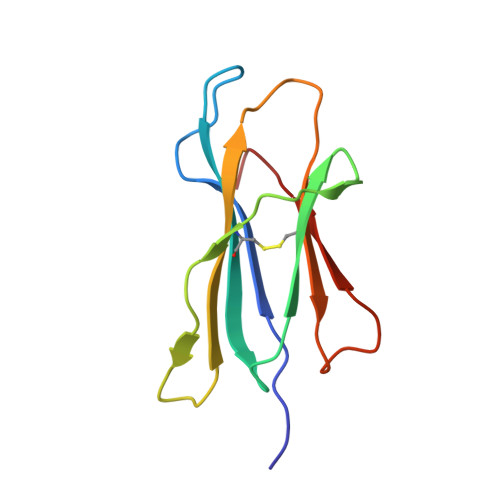
A Single T Cell Receptor Bound to Major Histocompatibility Complex Class I and Class II Glycoproteins Reveals Switchable TCR Conformers.
Yin, L., Huseby, E., Scott-Browne, J., Rubtsova, K., Pinilla, C., Crawford, F., Marrack, P., Dai, S., Kappler, J.W.(2011) Immunity 35: 23-33
- PubMed: 21683626
- DOI: https://doi.org/10.1016/j.immuni.2011.04.017
- Primary Citation of Related Structures:
3RGV - PubMed Abstract:
Major histocompatibility complex class I (MHCI) and MHCII proteins differ in structure and sequence. To understand how T cell receptors (TCRs) can use the same set of variable regions to bind both proteins, we have presented a comparison of a single TCR bound to both MHCI and MHCII ligands. The TCR adopts similar orientations on both ligands with TCR amino acids thought to be evolutionarily conserved for MHC interaction occupying similar positions on the MHCI and MHCII helices. However, the TCR antigen-binding loops use different conformations when interacting with each ligand. Most importantly, we observed alternate TCR core conformations. When bound to MHCI, but not MHCII, Vα disengages from the Jα β strand, switching Vα's position relative to Vβ. In several other structures, either Vα or Vβ undergoes this same modification. Thus, both TCR V-domains can switch among alternate conformations, perhaps extending their ability to react with different MHC-peptide ligands.
- Howard Hughes Medical Institute and Integrated Department of Immunology, National Jewish Health, Denver, CO 80206, USA.
Organizational Affiliation: