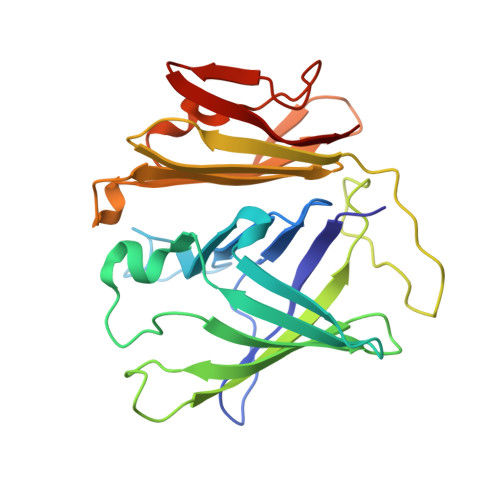
Novel beta-structure of YLR301w from Saccharomyces cerevisiae.
Kim, K.H., Ahn, H.J., Lee, W.K., Lee, C., Yu, M.H., Kim, E.E.(2012) Acta Crystallogr D Biol Crystallogr 68: 531-540
- PubMed: 22525751
- DOI: https://doi.org/10.1107/S090744491200491X
- Primary Citation of Related Structures:
3RBY - PubMed Abstract:
When the Z-type variant of human α(1)-antitrypsin was overexpressed in Saccharomyces cerevisiae, proteomics analysis identified YLR301w as one of the up-regulated proteins. YLR301w is a 27.5 kDa protein with no sequence homology to any known protein and has been reported to interact with Sec72 and Hrr25. The crystal structure of S. cerevisiae YLR301w has been determined at 2.3 Å resolution, revealing a novel β-structure. It consists of an N-terminal ten-stranded β-barrel with two short α-helices connected by a 23-residue linker to a seven-stranded half-barrel with two short helices at the C-terminus. The N-terminal barrel has a highly conserved hydrophobic channel that can bind hydrophobic molecules such as PEG. It forms a homodimer both in the crystal and in solution. YLR301w binds Sec72 with a K(d) of 6.2 µM, but the biological significance of this binding requires further investigation.
- Biomedical Research Institute, Korea Institute of Science and Technology, 39-1 Hawolkok-dong, Sungbuk-gu, Seoul 136-791, Republic of Korea.
Organizational Affiliation: