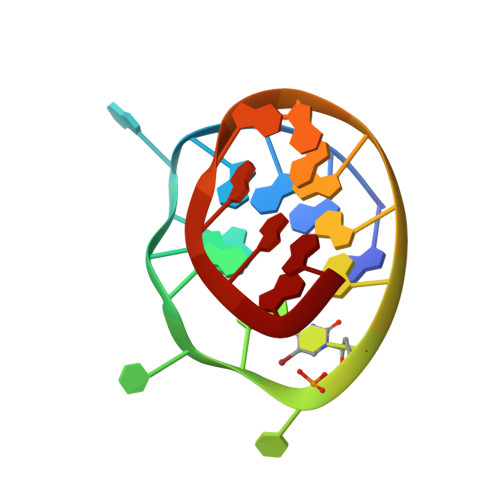
Crystal structure of a c-kit promoter quadruplex reveals the structural role of metal ions and water molecules in maintaining loop conformation.
Wei, D., Parkinson, G.N., Reszka, A.P., Neidle, S.(2012) Nucleic Acids Res 40: 4691-4700
- PubMed: 22287624
- DOI: https://doi.org/10.1093/nar/gks023
- Primary Citation of Related Structures:
3QXR - PubMed Abstract:
We report here the 1.62 Å crystal structure of an intramolecular quadruplex DNA formed from a sequence in the promoter region of the c-kit gene. This is the first reported crystal structure of a promoter quadruplex and the first observation of localized magnesium ions in a quadruplex structure. The structure reveals that potassium and magnesium ions have an unexpected yet significant structural role in stabilizing particular quadruplex loops and grooves that is distinct from but in addition to the role of potassium ions in the ion channel at the centre of all quadruplex structures. The analysis also shows how ions cluster together with structured water molecules to stabilize the quadruplex arrangement. This particular quadruplex has been previously studied by NMR methods, and the present X-ray structure is in accord with the earlier topology assignment. However, as well as the observations of potassium and magnesium ions, the crystal structure has revealed a highly significant difference in the dimensions of the large cleft in the structure, which is a plausible target for small molecules. This difference can be understood by the stabilizing role of structured water networks.
- CRUK Biomolecular Structure Group, UCL School of Pharmacy, University College London, 29-39 Brunswick Square, WC1N 1AX, London, UK.
Organizational Affiliation: