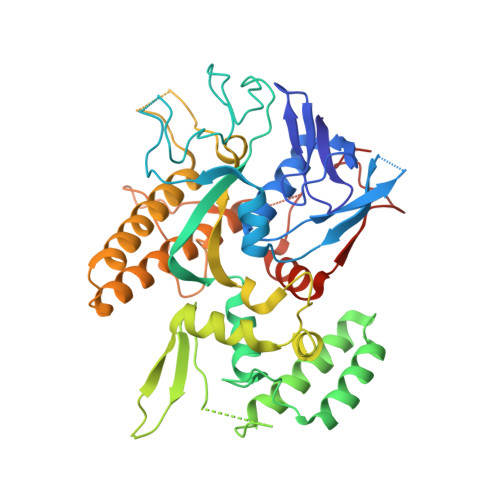
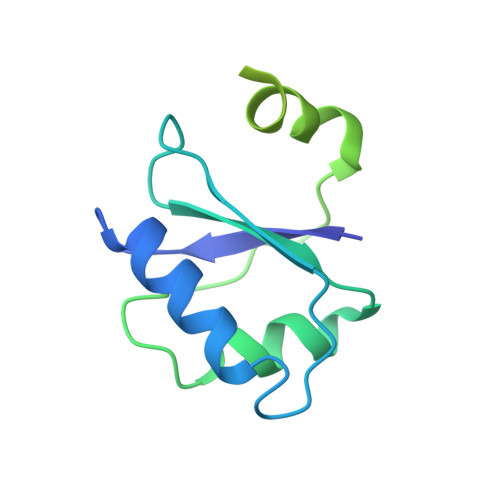
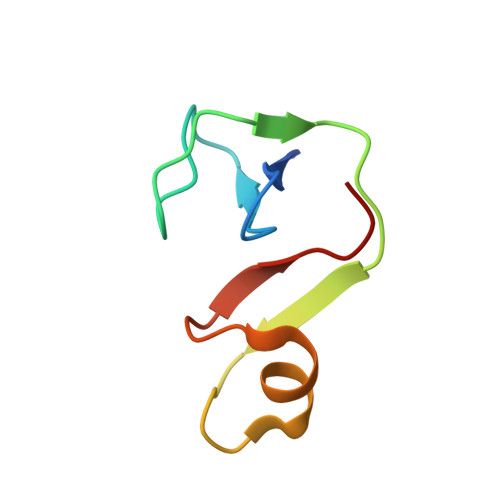
Architecture of the RNA polymerase-Spt4/5 complex and basis of universal transcription processivity.
Martinez-Rucobo, F.W., Sainsbury, S., Cheung, A.C., Cramer, P.(2011) EMBO J 30: 1302-1310
- PubMed: 21386817
- DOI: https://doi.org/10.1038/emboj.2011.64
- Primary Citation of Related Structures:
3QQC - PubMed Abstract:
Related RNA polymerases (RNAPs) carry out cellular gene transcription in all three kingdoms of life. The universal conservation of the transcription machinery extends to a single RNAP-associated factor, Spt5 (or NusG in bacteria), which renders RNAP processive and may have arisen early to permit evolution of long genes. Spt5 associates with Spt4 to form the Spt4/5 heterodimer. Here, we present the crystal structure of archaeal Spt4/5 bound to the RNAP clamp domain, which forms one side of the RNAP active centre cleft. The structure revealed a conserved Spt5-RNAP interface and enabled modelling of complexes of Spt4/5 counterparts with RNAPs from all kingdoms of life, and of the complete yeast RNAP II elongation complex with bound Spt4/5. The N-terminal NGN domain of Spt5/NusG closes the RNAP active centre cleft to lock nucleic acids and render the elongation complex stable and processive. The C-terminal KOW1 domain is mobile, but its location is restricted to a region between the RNAP clamp and wall above the RNA exit tunnel, where it may interact with RNA and/or other factors.
- Gene Center and Department of Biochemistry, Center for Integrated Protein Science Munich (CIPSM), Ludwig-Maximilians-Universität München, Munich, Germany.
Organizational Affiliation: