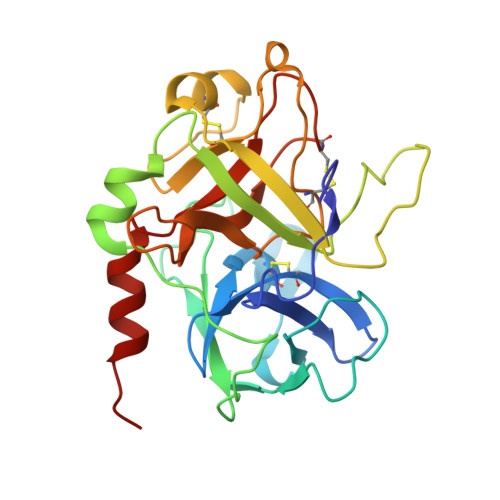
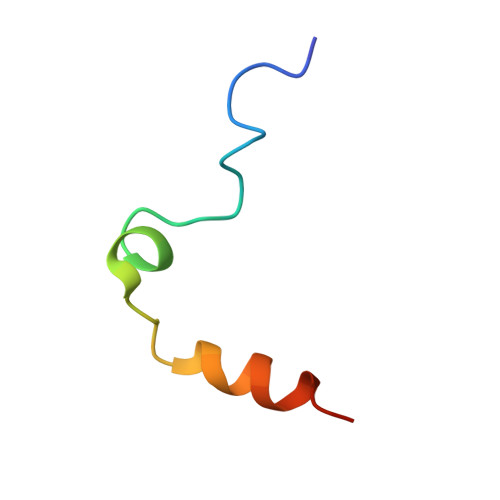
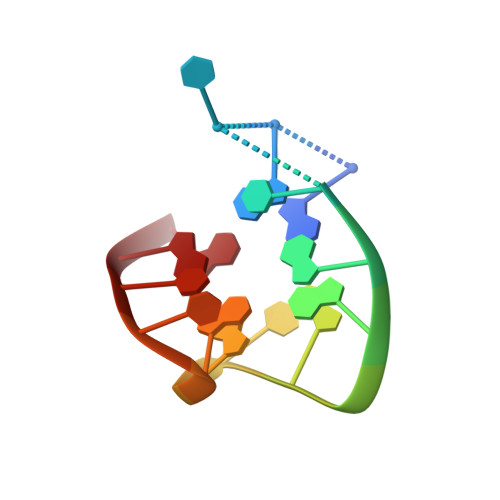
Thrombin-aptamer recognition: a revealed ambiguity.
Russo Krauss, I., Merlino, A., Giancola, C., Randazzo, A., Mazzarella, L., Sica, F.(2011) Nucleic Acids Res 39: 7858-7867
- PubMed: 21715374
- DOI: https://doi.org/10.1093/nar/gkr522
- Primary Citation of Related Structures:
3QLP - PubMed Abstract:
Aptamers are structured oligonucleotides that recognize molecular targets and can function as direct protein inhibitors. The best-known example is the thrombin-binding aptamer, TBA, a single-stranded 15-mer DNA that inhibits the activity of thrombin, the key enzyme of coagulation cascade. TBA folds as a G-quadruplex structure, as proved by its NMR structure. The X-ray structure of the complex between TBA and human α-thrombin was solved at 2.9-Å resolution, but did not provide details of the aptamer conformation and the interactions with the protein molecule. TBA is rapidly processed by nucleases. To improve the properties of TBA, a number of modified analogs have been produced. In particular, a modified TBA containing a 5'-5' polarity inversion site, mTBA, has higher stability and higher affinity toward thrombin with respect to TBA, although it has a lower inhibitory activity. We present the crystal structure of the thrombin-mTBA complex at 2.15-Å resolution; the resulting model eventually provides a clear picture of thrombin-aptamers interaction, and also highlights the structural bases of the different properties of TBA and mTBA. Our findings open the way for a rational design of modified aptamers with improved potency as anticoagulant drugs.
- Università di Napoli Federico II, Via Cintia, Napoli, Italia.
Organizational Affiliation: