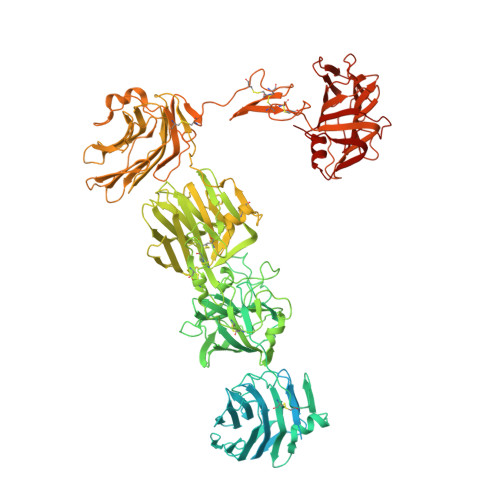
The Structure of Neurexin 1 alpha Reveals Features Promoting a Role as Synaptic Organizer
Chen, F., Venugopal, V., Murray, B., Rudenko, G.(2011) Structure 19: 779-789
- PubMed: 21620716
- DOI: https://doi.org/10.1016/j.str.2011.03.012
- Primary Citation of Related Structures:
3QCW, 3R05 - PubMed Abstract:
α-neurexins are essential synaptic adhesion molecules implicated in autism spectrum disorder and schizophrenia. The α-neurexin extracellular domain consists of six LNS domains interspersed by three EGF-like repeats and interacts with many different proteins in the synaptic cleft. To understand how α-neurexins might function as synaptic organizers, we solved the structure of the neurexin 1α extracellular domain (n1α) to 2.65 Å. The L-shaped molecule can be divided into a flexible repeat I (LNS1-EGF-A-LNS2), a rigid horseshoe-shaped repeat II (LNS3-EGF-B-LNS4) with structural similarity to so-called reelin repeats, and an extended repeat III (LNS5-EGF-B-LNS6) with controlled flexibility. A 2.95 Å structure of n1α carrying splice insert SS#3 in LNS4 reveals that SS#3 protrudes as a loop and does not alter the rigid arrangement of repeat II. The global architecture imposed by conserved structural features enables α-neurexins to recruit and organize proteins in distinct and variable ways, influenced by splicing, thereby promoting synaptic function.
- Life Sciences Institute, University of Michigan, Ann Arbor, MI 48109, USA.
Organizational Affiliation: