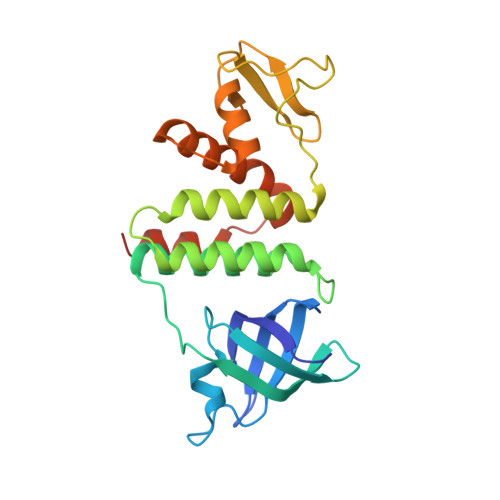
Mechanism of RecO recruitment to DNA by single-stranded DNA binding protein.
Ryzhikov, M., Koroleva, O., Postnov, D., Tran, A., Korolev, S.(2011) Nucleic Acids Res 39: 6305-6314
- PubMed: 21504984
- DOI: https://doi.org/10.1093/nar/gkr199
- Primary Citation of Related Structures:
3Q8D - PubMed Abstract:
RecO is a recombination mediator protein (RMP) important for homologous recombination, replication repair and DNA annealing in bacteria. In all pathways, the single-stranded (ss) DNA binding protein, SSB, plays an inhibitory role by protecting ssDNA from annealing and recombinase binding. Conversely, SSB may stimulate each reaction through direct interaction with RecO. We present a crystal structure of Escherichia coli RecO bound to the conserved SSB C-terminus (SSB-Ct). SSB-Ct binds the hydrophobic pocket of RecO in a conformation similar to that observed in the ExoI/SSB-Ct complex. Hydrophobic interactions facilitate binding of SSB-Ct to RecO and RecO/RecR complex in both low and moderate ionic strength solutions. In contrast, RecO interaction with DNA is inhibited by an elevated salt concentration. The SSB mutant lacking SSB-Ct also inhibits RecO-mediated DNA annealing activity in a salt-dependent manner. Neither RecO nor RecOR dissociates SSB from ssDNA. Therefore, in E. coli, SSB recruits RMPs to ssDNA through SSB-Ct, and RMPs are likely to alter the conformation of SSB-bound ssDNA without SSB dissociation to initiate annealing or recombination. Intriguingly, Deinococcus radiodurans RecO does not bind SSB-Ct and weakly interacts with the peptide in the presence of RecR, suggesting the diverse mechanisms of DNA repair pathways mediated by RecO in different organisms.
- Department of Biochemistry and Molecular Biology, St Louis University School of Medicine, 1100 S Grand Blvd, St Louis, MO 63021, USA.
Organizational Affiliation: