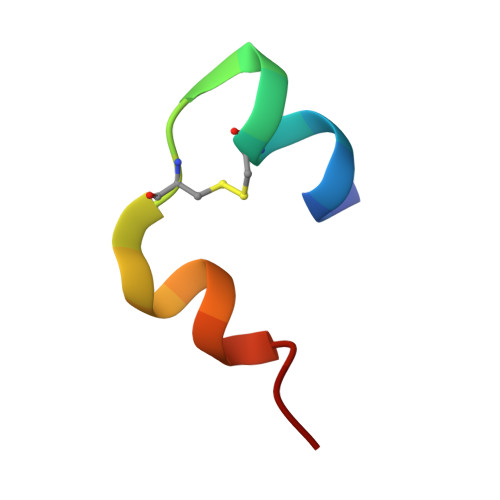
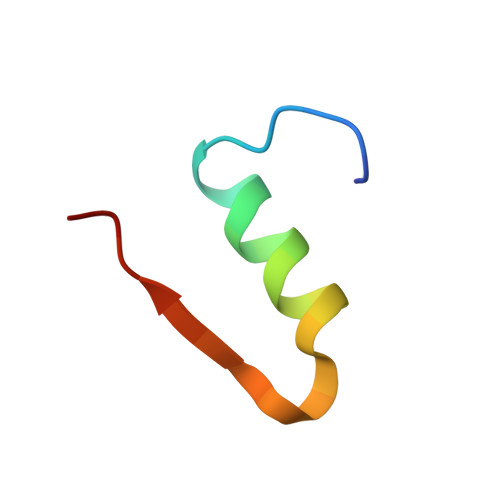
Molecular recognition of insulin by a synthetic receptor.
Chinai, J.M., Taylor, A.B., Ryno, L.M., Hargreaves, N.D., Morris, C.A., Hart, P.J., Urbach, A.R.(2011) J Am Chem Soc 133: 8810-8813
- PubMed: 21473587
- DOI: https://doi.org/10.1021/ja201581x
- Primary Citation of Related Structures:
3Q6E - PubMed Abstract:
The discovery of molecules that bind tightly and selectively to desired proteins continues to drive innovation at the interface of chemistry and biology. This paper describes the binding of human insulin by the synthetic receptor cucurbit[7]uril (Q7) in vitro. Isothermal titration calorimetry and fluorescence spectroscopy experiments show that Q7 binds to insulin with an equilibrium association constant of 1.5 × 10(6) M(-1) and with 50-100-fold selectivity versus proteins that are much larger but lack an N-terminal aromatic residue, and with >1000-fold selectivity versus an insulin variant lacking the N-terminal phenylalanine (Phe) residue. The crystal structure of the Q7·insulin complex shows that binding occurs at the N-terminal Phe residue and that the N-terminus unfolds to enable binding. These findings suggest that site-selective recognition is based on the properties inherent to a protein terminus, including the unique chemical epitope presented by the terminal residue and the greater freedom of the terminus to unfold, like the end of a ball of string, to accommodate binding. Insulin recognition was predicted accurately from studies on short peptides and exemplifies an approach to protein recognition by targeting the terminus.
- Department of Chemistry, Trinity University, One Trinity Place, San Antonio, Texas 78212, USA.
Organizational Affiliation: