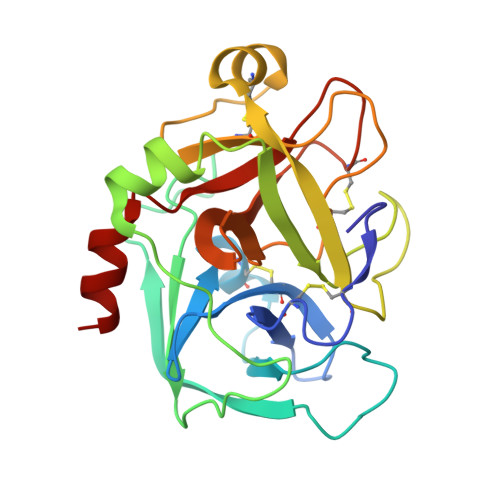
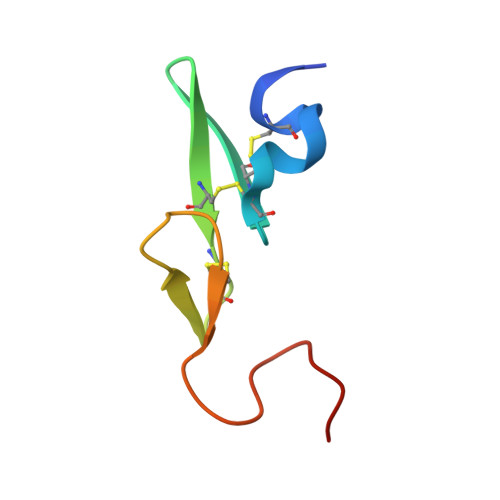
Design, synthesis and SAR of novel ethylenediamine and phenylenediamine derivatives as factor Xa inhibitors.
Yoshikawa, K., Yoshino, T., Yokomizo, Y., Uoto, K., Naito, H., Kawakami, K., Mochizuki, A., Nagata, T., Suzuki, M., Kanno, H., Takemura, M., Ohta, T.(2011) Bioorg Med Chem Lett 21: 2133-2140
- PubMed: 21345673
- DOI: https://doi.org/10.1016/j.bmcl.2011.01.132
- Primary Citation of Related Structures:
3Q3K - PubMed Abstract:
We previously reported on a series of cyclohexanediamine derivatives as highly potent factor Xa inhibitors. Herein, we describe the modification of the spacer moiety to discover an alternative scaffold. Ethylenediamine derivatives possessing a substituent at the C1 position in S configuration and phenylenediamine derivatives possessing a substituent at the C5 position demonstrated moderate to strong anti-fXa activity. Further SAR studies led to the identification of compound 30 h which showed both good in vitro activity (fXa IC(50) = 2.2 nM, PTCT2 = 3.9 μM) and in vivo antithrombotic efficacy.
- R&D Division, Daiichi Sankyo Co, Ltd, 1-16-13, Kita-Kasai, Edogawa-ku, Tokyo 134-8630, Japan. yoshikawa.kenji.t6@daiichisankyo.co.jp
Organizational Affiliation: