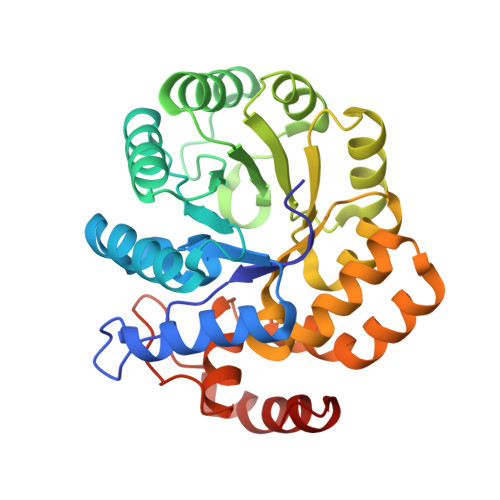
Crystal structure of the complex of Dhydrodipicolinate synthase from Acinetobacter baumannii with lysine at 2.6A resolution
Jithesh, O., Yamini, S., Kaur, N., Gautam, A., Tewari, R., Kushwaha, G.S., Kaur, P., Srinivasan, A., Sharma, S., Singh, T.P.To be published.