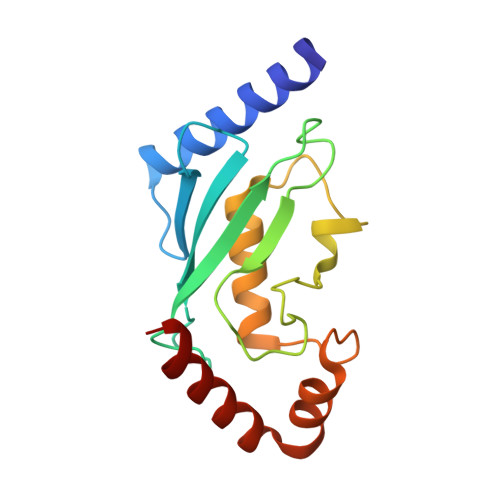
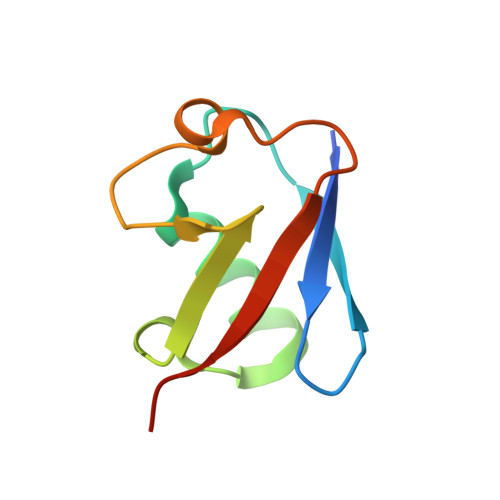
Modulation of K11-Linkage Formation by Variable Loop Residues within UbcH5A.
Bosanac, I., Phu, L., Pan, B., Zilberleyb, I., Maurer, B., Dixit, V.M., Hymowitz, S.G., Kirkpatrick, D.S.(2011) J Mol Biology 408: 420-431
- PubMed: 21396940
- DOI: https://doi.org/10.1016/j.jmb.2011.03.011
- Primary Citation of Related Structures:
3PTF - PubMed Abstract:
Ubiquitination refers to the covalent addition of ubiquitin (Ub) to substrate proteins or other Ub molecules via the sequential action of three enzymes (E1, E2, and E3). Recent advances in mass spectrometry proteomics have made it possible to identify and quantify Ub linkages in biochemical and cellular systems. We used these tools to probe the mechanisms controlling linkage specificity for UbcH5A. UbcH5A is a promiscuous E2 enzyme with an innate preference for forming polyubiquitin chains through lysine 11 (K11), lysine 48 (K48), and lysine 63 (K63) of Ub. We present the crystal structure of a noncovalent complex between Ub and UbcH5A. This structure reveals an interaction between the Ub surface flanking K11 and residues adjacent to the E2 catalytic cysteine and suggests a possible role for this surface in formation of K11 linkages. Structure-guided mutagenesis, in vitro ubiquitination and quantitative mass spectrometry have been used to characterize the ability of residues in the vicinity of the E2 active site to direct synthesis of K11- and K63-linked polyubiquitin. Mutation of critical residues in the interface modulated the linkage specificity of UbcH5A, resulting in generation of more K63-linked chains at the expense of K11-linkage synthesis. This study provides direct evidence that the linkage specificity of E2 enzymes may be altered through active-site mutagenesis.
- Department of Structural Biology, Genentech Inc., 1 DNA Way, South San Francisco, CA 94080, USA.
Organizational Affiliation: