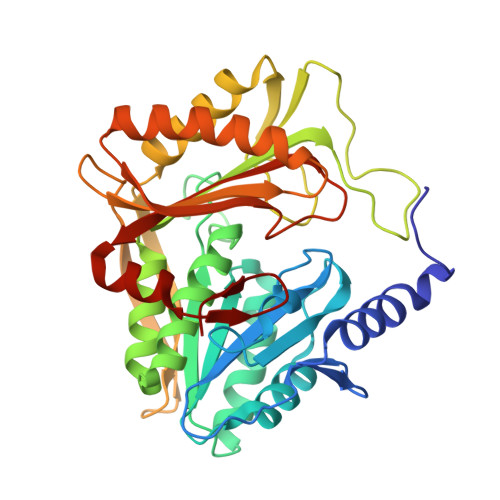
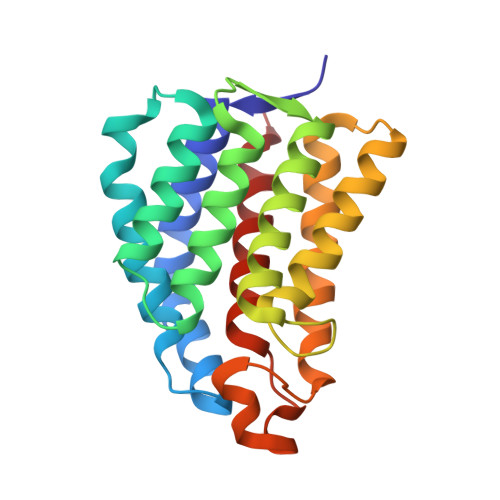
Structural and mechanistic insight into covalent substrate binding by Escherichia coli dihydroxyacetone kinase.
Shi, R., McDonald, L., Cui, Q., Matte, A., Cygler, M., Ekiel, I.(2011) Proc Natl Acad Sci U S A 108: 1302-1307
- PubMed: 21209328
- DOI: https://doi.org/10.1073/pnas.1012596108
- Primary Citation of Related Structures:
3PNK, 3PNL, 3PNM, 3PNO, 3PNQ - PubMed Abstract:
The Escherichia coli dihydroxyacetone (Dha) kinase is an unusual kinase because (i) it uses the phosphoenolpyruvate carbohydrate: phosphotransferase system (PTS) as the source of high-energy phosphate, (ii) the active site is formed by two subunits, and (iii) the substrate is covalently bound to His218(K)* of the DhaK subunit. The PTS transfers phosphate to DhaM, which in turn phosphorylates the permanently bound ADP coenzyme of DhaL. This phosphoryl group is subsequently transferred to the Dha substrate bound to DhaK. Here we report the crystal structure of the E. coli Dha kinase complex, DhaK-DhaL. The structure of the complex reveals that DhaK undergoes significant conformational changes to accommodate binding of DhaL. Combined mutagenesis and enzymatic activity studies of kinase mutants allow us to propose a catalytic mechanism for covalent Dha binding, phosphorylation, and release of the Dha-phosphate product. Our results show that His56(K) is involved in formation of the covalent hemiaminal bond with Dha. The structure of H56N(K) with noncovalently bound substrate reveals a somewhat different positioning of Dha in the binding pocket as compared to covalently bound Dha, showing that the covalent attachment to His218(K) orients the substrate optimally for phosphoryl transfer. Asp109(K) is critical for activity, likely acting as a general base activating the γ-OH of Dha. Our results provide a comprehensive picture of the roles of the highly conserved active site residues of dihydroxyacetone kinases.
- Department of Biochemistry, McGill University, Montréal, QC, Canada H3G 1Y6.
Organizational Affiliation: