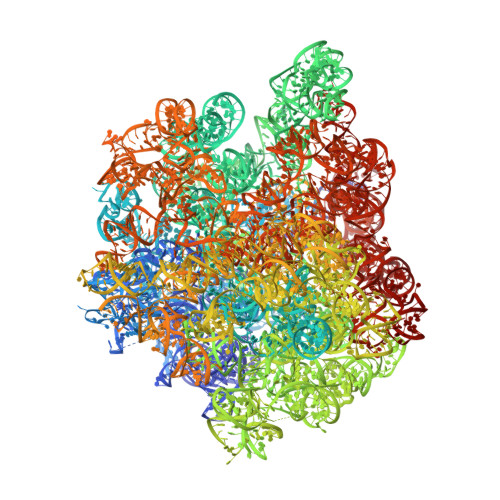
Crystal structure of the synergistic antibiotic pair, lankamycin and lankacidin, in complex with the large ribosomal subunit.
Belousoff, M.J., Shapira, T., Bashan, A., Zimmerman, E., Rozenberg, H., Arakawa, K., Kinashi, H., Yonath, A.(2011) Proc Natl Acad Sci U S A 108: 2717-2722
- PubMed: 21282615
- DOI: https://doi.org/10.1073/pnas.1019406108
- Primary Citation of Related Structures:
3PIO, 3PIP - PubMed Abstract:
The structures of the large ribosomal subunit of Deinococcus radiodurans (D50S) in complex with the antibiotic lankamycin (3.2 Å) and a double antibiotic complex of lankamycin and lankacidin C (3.45 Å) have been determined, in continuation of previous crystallographic studies on lankacidin-D50S complex. These two drugs have been previously reported to inhibit ribosomal function with mild synergistic effect. Lankamycin, a member of the macrolide family, binds in a similar manner to erythromycin. However, when in complex with lankacidin, lankamycin is located so that it can form interactions with lankacidin in the adjacent ribosomal binding site. When compared to the well-documented synergistic antibiotics, Streptogramins A and B, the pair of lankacidin and lankamycin bind in similar sites, the peptidyl transferase center and nascent peptide exit tunnel, respectively. Herein, we discuss the structural basis for antibiotic synergism and highlight the key factors involved in ribosomal inhibition.
- Department of Structural Biology, Weizmann Institute of Science, Rehovot, 76100, Israel.
Organizational Affiliation: