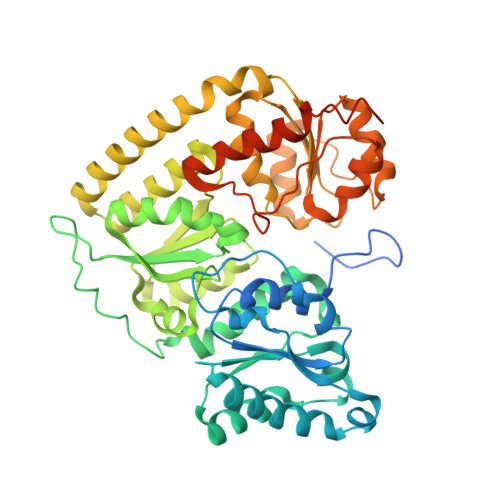
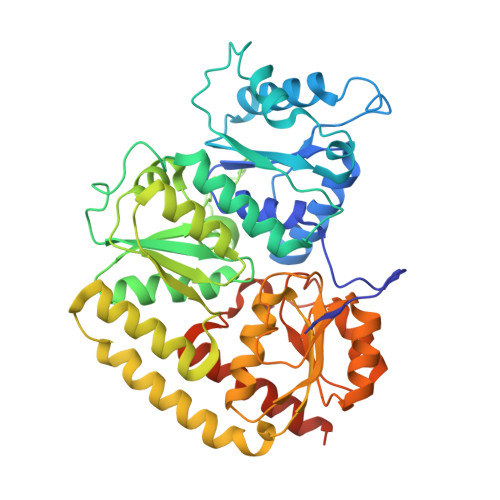
Structure of Precursor-Bound NifEN: A Nitrogenase FeMo Cofactor Maturase/Insertase.
Kaiser, J.T., Hu, Y., Wiig, J.A., Rees, D.C., Ribbe, M.W.(2011) Science 331: 91-94
- PubMed: 21212358
- DOI: https://doi.org/10.1126/science.1196954
- Primary Citation of Related Structures:
3PDI - PubMed Abstract:
NifEN plays an essential role in the biosynthesis of the nitrogenase iron-molybdenum (FeMo) cofactor (M cluster). It is an α(2)β(2) tetramer that is homologous to the catalytic molybdenum-iron (MoFe) protein (NifDK) component of nitrogenase. NifEN serves as a scaffold for the conversion of an iron-only precursor to a matured form of the M cluster before delivering the latter to its target location within NifDK. Here, we present the structure of the precursor-bound NifEN of Azotobacter vinelandii at 2.6 angstrom resolution. From a structural comparison of NifEN with des-M-cluster NifDK and holo NifDK, we propose similar pathways of cluster insertion for the homologous NifEN and NifDK proteins.
- Division of Chemistry and Chemical Engineering, California Institute of Technology, Mail Code 114-96, Pasadena, CA 91125, USA.
Organizational Affiliation: