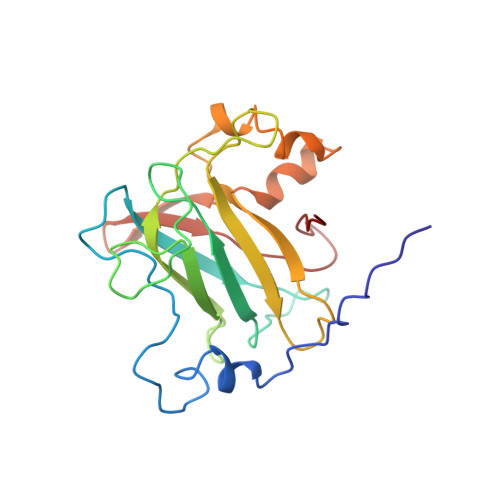
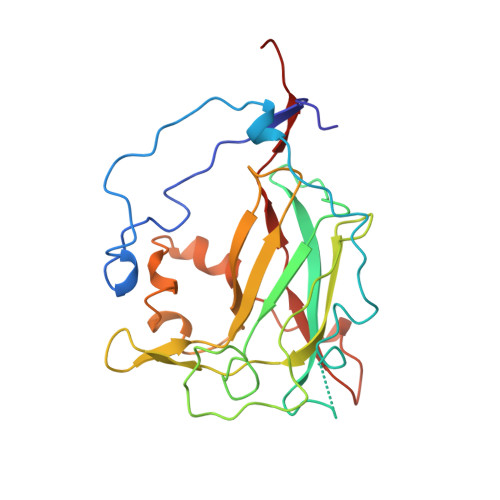
Crystal structure and resonance Raman studies of protocatechuate 3,4-dioxygenase complexed with 3,4-dihydroxyphenylacetate.
Elgren, T.E., Orville, A.M., Kelly, K.A., Lipscomb, J.D., Ohlendorf, D.H., Que Jr., L.(1997) Biochemistry 36: 11504-11513
- PubMed: 9298971
- DOI: https://doi.org/10.1021/bi970691k
- Primary Citation of Related Structures:
3PCN - PubMed Abstract:
The crystal structure of the anaerobic complex of Pseudomonas putida protocatechuate 3,4-dioxygenase (3,4-PCD) bound with the alternative substrate, 3,4-dihydroxyphenylacetate (HPCA), is reported at 2.4 A resolution and refined to an R factor of 0.17. Formation of the active site Fe(III).HPCA chelated complex causes the endogenous axial tyrosinate, Tyr447 (147beta), to dissociate from the iron and rotate into an alternative orientation analogous to that previously observed in the anaerobic 3,4-PCD.3,4-dihydroxybenzoate complex (3, 4-PCD.PCA) [Orville, A. M., Lipscomb, J. D., & Ohlendorf, D. H. (1997) Biochemistry 36, 10052-10066]. Two orientations of the aromatic ring of HPCA related by an approximate 180 degrees rotation within the active site are consistent with the electron density. Resonance Raman (rR) spectroscopic data from Brevibacteriumfuscum 3,4-PCD.HPCA complex in solution reveals low frequency rR vibrational bands between 500 and 650 cm-1 as well as a band at approximately 1320 cm-1 which are diagnostic of a HPCA. Fe(III) chelate complex. 18O labeling of HPCA at either the C4 or C3 hydroxyl group unambiguously establishes the vibrational coupling modes associated with the five-membered chelate ring system. Analysis of these data suggests that the Fe(III)-HPCAO4 bond is shorter than the Fe(III)-HPCAO3 bond. This consequently favors the model for the crystal structure in which the C3 phenolic function occupies the Fe3+ ligand site opposite the endogenous ligand Tyr408(Oeta) (108beta). This is essentially the same binding orientation as proposed for PCA in the crystal structure of the anaerobic 3,4-PCD.PCA complex based solely on direct modeling of the 2Fo - Fc electron density and suggests that this is the conformation required for catalysis.
- Department of Chemistry, Hamilton College, Clinton, New York 13323, USA.
Organizational Affiliation: