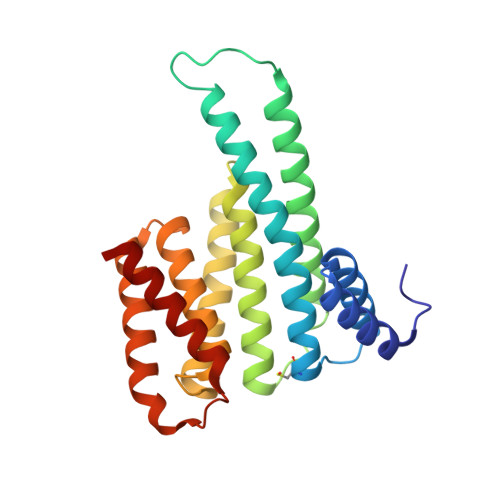
A semisynthetic fusicoccane stabilizes a protein-protein interaction and enhances the expression of K+ channels at the cell surface
Anders, C., Higuchi, Y., Koschinsky, K., Bartel, M., Schumacher, B., Thiel, P., Nitta, H., Preisig-Muller, R., Schlichthorl, G., Renigunta, V., Ohkanda, J., Daut, J., Kato, N., Ottmann, C.(2013) Chem Biol 20: 583-593
- PubMed: 23601647
- DOI: https://doi.org/10.1016/j.chembiol.2013.03.015
- Primary Citation of Related Structures:
3P1N, 3P1O, 3P1P, 3P1Q, 3P1R, 3P1S, 3SMK, 3SML, 3SMM, 3SMN, 3SMO, 3SP5, 3SPR, 3UX0, 4FR3 - PubMed Abstract:
Small-molecule stabilization of protein-protein interactions is an emerging field in chemical biology. We show how fusicoccanes, originally identified as fungal toxins acting on plants, promote the interaction of 14-3-3 proteins with the human potassium channel TASK-3 and present a semisynthetic fusicoccane derivative (FC-THF) that targets the 14-3-3 recognition motif (mode 3) in TASK-3. In the presence of FC-THF, the binding of 14-3-3 proteins to TASK-3 was increased 19-fold and protein crystallography provided the atomic details of the effects of FC-THF on this interaction. We also tested the functional effects of FC-THF on TASK channels heterologously expressed in Xenopus oocytes. Incubation with 10 μM FC-THF was found to promote the transport of TASK channels to the cell membrane, leading to a significantly higher density of channels at the surface membrane and increased potassium current.
- Chemical Genomics Centre of the Max-Planck-Society, 44227 Dortmund, Germany.
Organizational Affiliation: