Ribosome Recycling in Archaea A Mechanistic Link between FeS Cluster Biosynthesis and Conformational Switch of Twin-ATPase ABCE1
Barthelme, D., Dinkelaker, S., Albers, S.-V., Londei, P., Ermler, U., Tampe, R.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| RNase L inhibitor | 538 | Saccharolobus solfataricus | Mutation(s): 2 Gene Names: SSO0287 | 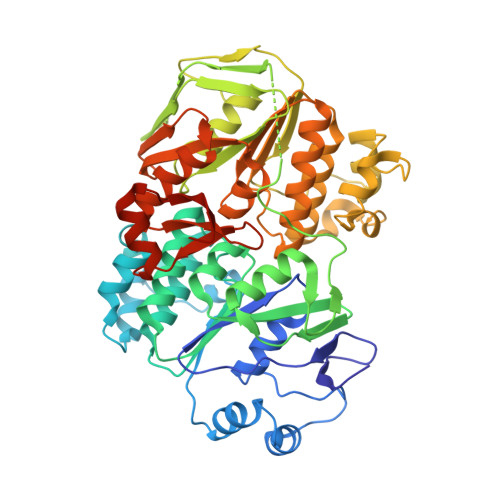 | |
UniProt | |||||
Find proteins for Q980K5 (Saccharolobus solfataricus (strain ATCC 35092 / DSM 1617 / JCM 11322 / P2)) Explore Q980K5 Go to UniProtKB: Q980K5 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q980K5 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| ADP Query on ADP | E [auth A], F [auth A], J [auth B], K [auth B] | ADENOSINE-5'-DIPHOSPHATE C10 H15 N5 O10 P2 XTWYTFMLZFPYCI-KQYNXXCUSA-N |  | ||
| PO4 Query on PO4 | G [auth A] | PHOSPHATE ION O4 P NBIIXXVUZAFLBC-UHFFFAOYSA-K |  | ||
| MG Query on MG | C [auth A], D [auth A], H [auth B], I [auth B] | MAGNESIUM ION Mg JLVVSXFLKOJNIY-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 58.374 | α = 89.15 |
| b = 63.656 | β = 84.47 |
| c = 81.778 | γ = 69.82 |
| Software Name | Purpose |
|---|---|
| MAR345dtb | data collection |
| PHASER | phasing |
| REFMAC | refinement |
| XDS | data reduction |
| XSCALE | data scaling |