Multiple C-terminal tail Ca(2+)/CaMs regulate Ca(V)1.2 function but do not mediate channel dimerization.
Kim, E.Y., Rumpf, C.H., Van Petegem, F., Arant, R.J., Findeisen, F., Cooley, E.S., Isacoff, E.Y., Minor, D.L.(2010) EMBO J 29: 3924-3938
- PubMed: 20953164
- DOI: https://doi.org/10.1038/emboj.2010.260
- Primary Citation of Related Structures:
3OXQ - PubMed Abstract:
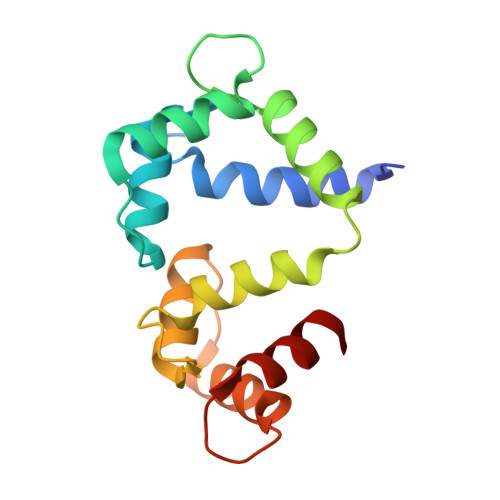
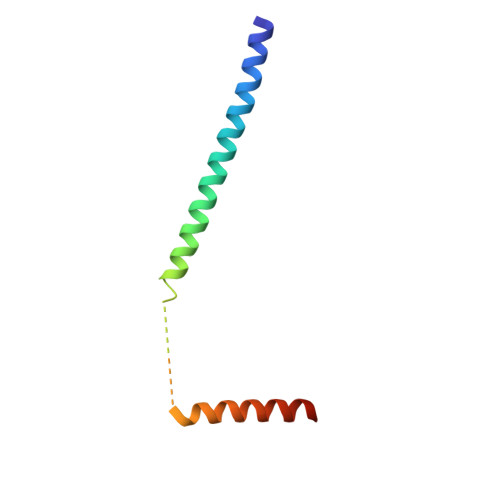
Interactions between voltage-gated calcium channels (Ca(V)s) and calmodulin (CaM) modulate Ca(V) function. In this study, we report the structure of a Ca(2+)/CaM Ca(V)1.2 C-terminal tail complex that contains two PreIQ helices bridged by two Ca(2+)/CaMs and two Ca(2+)/CaM-IQ domain complexes. Sedimentation equilibrium experiments establish that the complex has a 2:1 Ca(2+)/CaM:C-terminal tail stoichiometry and does not form higher order assemblies. Moreover, subunit-counting experiments demonstrate that in live cell membranes Ca(V)1.2s are monomers. Thus, contrary to previous proposals, the crystallographic dimer lacks physiological relevance. Isothermal titration calorimetry and biochemical experiments show that the two Ca(2+)/CaMs in the complex have different properties. Ca(2+)/CaM bound to the PreIQ C-region is labile, whereas Ca(2+)/CaM bound to the IQ domain is not. Furthermore, neither of lobes of apo-CaM interacts strongly with the PreIQ domain. Electrophysiological studies indicate that the PreIQ C-region has a role in calcium-dependent facilitation. Together, the data show that two Ca(2+)/CaMs can bind the Ca(V)1.2 tail simultaneously and indicate a functional role for Ca(2+)/CaM at the C-region site.
- Cardiovascular Research Institute, University of California, San Francisco, CA 94158-2330, USA.
Organizational Affiliation: