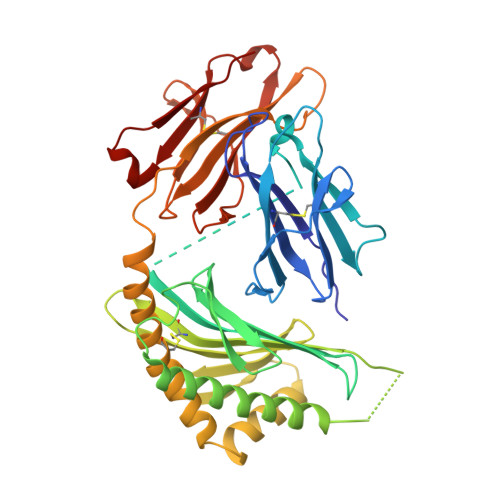
The 2.5 A structure of CD1c in complex with a mycobacterial lipid reveals an open groove ideally suited for diverse antigen presentation
Scharf, L., Li, N.S., Hawk, A.J., Garzon, D., Zhang, T., Fox, L.M., Kazen, A.R., Shah, S., Haddadian, E.J., Gumperz, J.E., Saghatelian, A., Faraldo-Gomez, J.D., Meredith, S.C., Piccirilli, J.A., Adams, E.J.(2010) Immunity 33: 853-862
- PubMed: 21167756
- DOI: https://doi.org/10.1016/j.immuni.2010.11.026
- Primary Citation of Related Structures:
3OV6 - PubMed Abstract:
CD1 molecules function to present lipid-based antigens to T cells. Here we present the crystal structure of CD1c at 2.5 Å resolution, in complex with the pathogenic Mycobacterium tuberculosis antigen mannosyl-β1-phosphomycoketide (MPM). CD1c accommodated MPM's methylated alkyl chain exclusively in the A' pocket, aided by a unique exit portal underneath the α1 helix. Most striking was an open F' pocket architecture lacking the closed cavity structure of other CD1 molecules, reminiscent of peptide binding grooves of classical major histocompatibility complex molecules. This feature, combined with tryptophan-fluorescence quenching during loading of a dodecameric lipopeptide antigen, provides a compelling model by which both the lipid and peptide moieties of the lipopeptide are involved in CD1c presentation of lipopeptides.
- Department of Biochemistry and Molecular Biology, University of Chicago, Chicago, IL 60637, USA.
Organizational Affiliation: