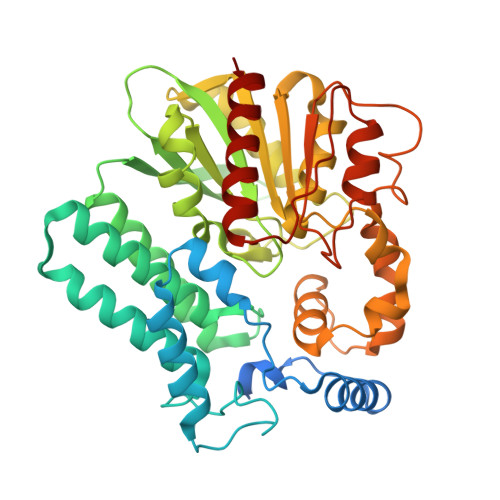
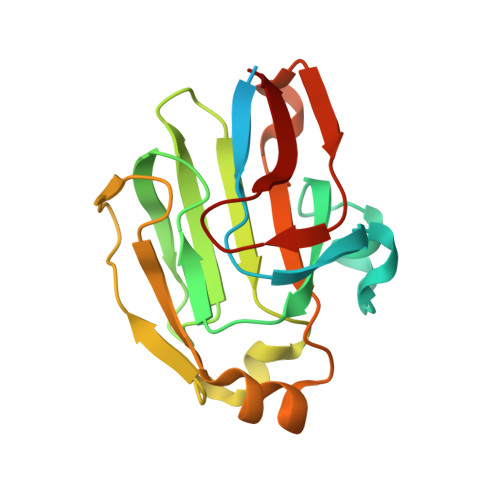
FrsA functions as a cofactor-independent decarboxylase to control metabolic flux
Lee, K.J., Jeong, C.S., An, Y.J., Lee, H.J., Park, S.J., Seok, Y.J., Kim, P., Lee, J.H., Lee, K.H., Cha, S.S.(2011) Nat Chem Biol 7: 434-436
- PubMed: 21623357
- DOI: https://doi.org/10.1038/nchembio.589
- Primary Citation of Related Structures:
3MVE, 3OUR - PubMed Abstract:
The interaction between fermentation-respiration switch (FrsA) protein and glucose-specific enzyme IIA(Glc) increases glucose fermentation under oxygen-limited conditions. We show that FrsA converts pyruvate to acetaldehyde and carbon dioxide in a cofactor-independent manner and that its pyruvate decarboxylation activity is enhanced by the dephosphorylated form of IIA(Glc) (d-IIA(Glc)). Crystal structures of FrsA and its complex with d-IIA(Glc) revealed residues required for catalysis as well as the structural basis for the activation by d-IIA(Glc).
- Department of Environmental Science, Hankuk University of Foreign Studies, Yongin, Republic of Korea.
Organizational Affiliation: