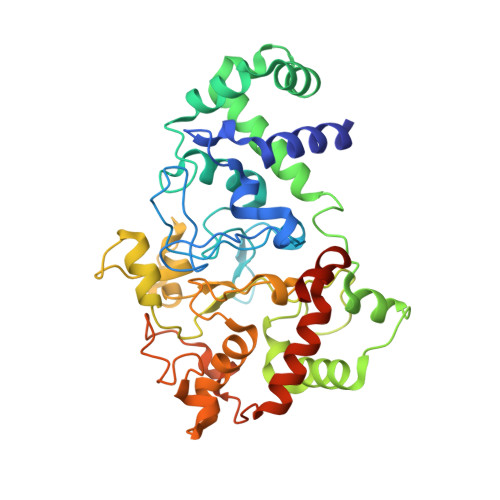
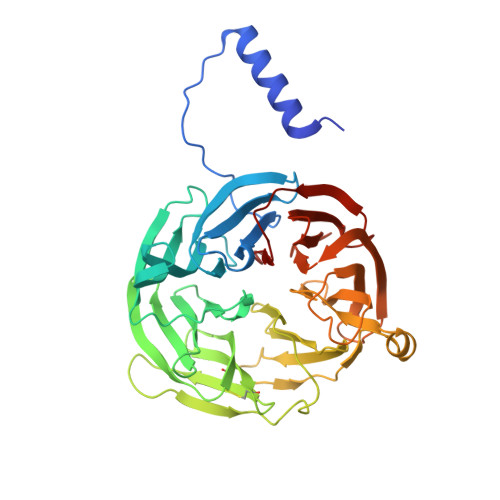
Functional Importance of Tyrosine 294 and the Catalytic Selectivity for the Bis-Fe(IV) State of MauG Revealed by Replacement of This Axial Heme Ligand with Histidine .
Abu Tarboush, N., Jensen, L.M., Feng, M., Tachikawa, H., Wilmot, C.M., Davidson, V.L.(2010) Biochemistry 49: 9783-9791
- PubMed: 20929212
- DOI: https://doi.org/10.1021/bi101254p
- Primary Citation of Related Structures:
3ORV - PubMed Abstract:
The diheme enzyme MauG catalyzes the posttranslational modification of a precursor protein of methylamine dehydrogenase (preMADH) to complete the biosynthesis of its protein-derived tryptophan tryptophylquinone (TTQ) cofactor. It catalyzes three sequential two-electron oxidation reactions which proceed through a high-valent bis-Fe(IV) redox state. Tyr294, the unusual distal axial ligand of one c-type heme, was mutated to His, and the crystal structure of Y294H MauG in complex with preMADH reveals that this heme now has His-His axial ligation. Y294H MauG is able to interact with preMADH and participate in interprotein electron transfer, but it is unable to catalyze the TTQ biosynthesis reactions that require the bis-Fe(IV) state. This mutation affects not only the redox properties of the six-coordinate heme but also the redox and CO-binding properties of the five-coordinate heme, despite the 21 Å separation of the heme iron centers. This highlights the communication between the hemes which in wild-type MauG behave as a single diheme unit. Spectroscopic data suggest that Y294H MauG can stabilize a high-valent redox state equivalent to Fe(V), but it appears to be an Fe(IV)═O/π radical at the five-coordinate heme rather than the bis-Fe(IV) state. This compound I-like intermediate does not catalyze TTQ biosynthesis, demonstrating that the bis-Fe(IV) state, which is stabilized by Tyr294, is specifically required for this reaction. The TTQ biosynthetic reactions catalyzed by wild-type MauG do not occur via direct contact with the Fe(IV)═O heme but via long-range electron transfer through the six-coordinate heme. Thus, a critical feature of the bis-Fe(IV) species may be that it shortens the electron transfer distance from preMADH to a high-valent heme iron.
- Department of Biochemistry, University of Mississippi Medical Center, Jackson, Mississippi 39216, United States.
Organizational Affiliation: