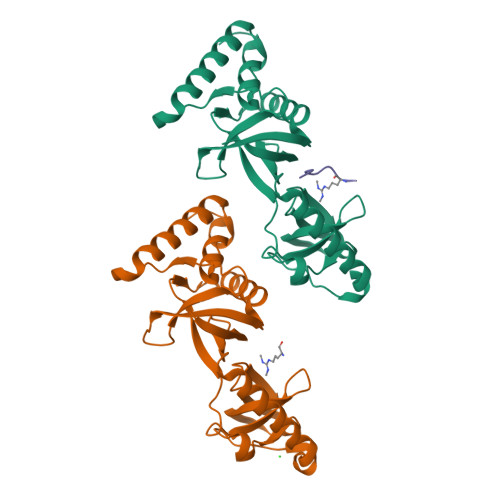
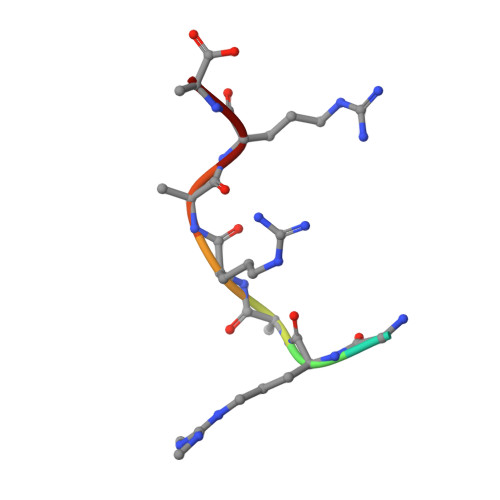
Structural basis for recognition of arginine methylated Piwi proteins by the extended Tudor domain.
Liu, K., Chen, C., Guo, Y., Lam, R., Bian, C., Xu, C., Zhao, D.Y., Jin, J., MacKenzie, F., Pawson, T., Min, J.(2010) Proc Natl Acad Sci U S A 107: 18398-18403
- PubMed: 20937909
- DOI: https://doi.org/10.1073/pnas.1013106107
- Primary Citation of Related Structures:
3OMC, 3OMG - PubMed Abstract:
Arginine methylation modulates diverse cellular processes and represents a molecular signature of germ-line-specific Piwi family proteins. A subset of Tudor domains recognize arginine methylation modifications, but the binding mechanism has been lacking. Here we establish that, like other germ-line Tudor proteins, the ancestral staphylococcal nuclease domain-containing 1 (SND1) polypeptide is expressed and associates with PIWIL1/Miwi in germ cells. We find that human SND1 binds PIWIL1 in an arginine methylation-dependent manner with a preference for symmetrically dimethylated arginine. The entire Tudor domain and a bifurcated SN domain are required for this binding activity, whereas the canonical Tudor domain alone is insufficient for methylarginine ligand binding. Crystal structures show that the intact SND1 extended Tudor domain forms a wide and negatively charged binding groove, which can accommodate distinct symmetrically dimethylated arginine peptides from PIWIL1 in different orientations. This analysis explains how SND1 preferentially recognizes symmetrical dimethylarginine via an aromatic cage and conserved hydrogen bonds, and provides a general paradigm for the binding mechanisms of methylarginine-containing peptides by extended Tudor domains.
Organizational Affiliation:
Hubei Key Laboratory of Genetic Regulation and Integrative Biology, College of Life Science, Huazhong Normal University, Wuhan 430079, People's Republic of China.