Structural basis for error-free replication of oxidatively damaged DNA by yeast DNA polymerase eta.
Silverstein, T.D., Jain, R., Johnson, R.E., Prakash, L., Prakash, S., Aggarwal, A.K.(2010) Structure 18: 1463-1470
- PubMed: 21070945
- DOI: https://doi.org/10.1016/j.str.2010.08.019
- Primary Citation of Related Structures:
3OHA, 3OHB - PubMed Abstract:
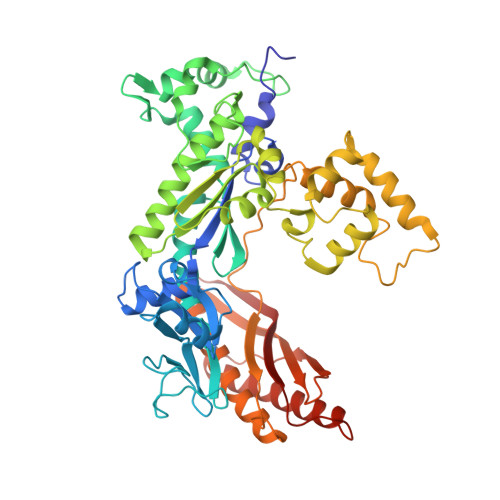
7,8-dihydro-8-oxoguanine (8-oxoG) adducts are formed frequently by the attack of oxygen-free radicals on DNA. They are among the most mutagenic lesions in cells because of their dual coding potential, where, in addition to normal base-pairing of 8-oxoG(anti) with dCTP, 8-oxoG in the syn conformation can base pair with dATP, causing G to T transversions. We provide here for the first time a structural basis for the error-free replication of 8-oxoG lesions by yeast DNA polymerase η (Polη). We show that the open active site cleft of Polη can accommodate an 8-oxoG lesion in the anti conformation with only minimal changes to the polymerase and the bound DNA: at both the insertion and post-insertion steps of lesion bypass. Importantly, the active site geometry remains the same as in the undamaged complex and provides a basis for the ability of Polη to prevent the mutagenic replication of 8-oxoG lesions in cells.
- Department of Structural & Chemical Biology, Mount Sinai School of Medicine, Box 1677, 1425 Madison Avenue, New York, NY 10029, USA.
Organizational Affiliation: