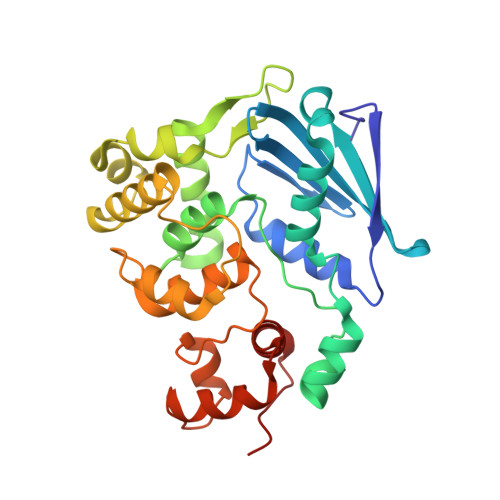
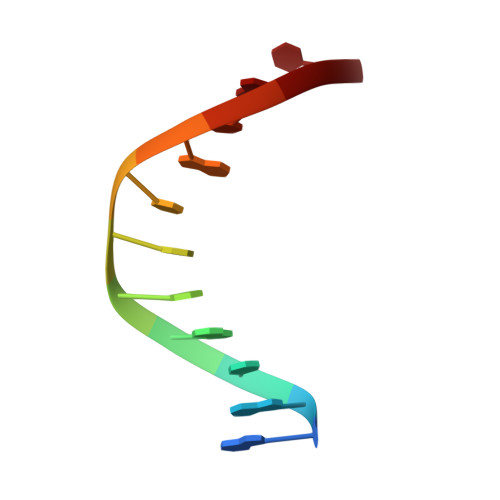
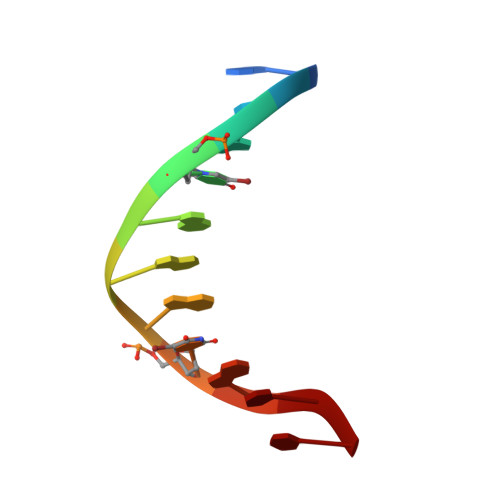
Structure of Escherichia coli AlkA in Complex with Undamaged DNA.
Bowman, B.R., Lee, S., Wang, S., Verdine, G.L.(2010) J Biological Chem 285: 35783-35791
- PubMed: 20843803
- DOI: https://doi.org/10.1074/jbc.M110.155663
- Primary Citation of Related Structures:
3OGD, 3OH6, 3OH9 - PubMed Abstract:
Because DNA damage is so rare, DNA glycosylases interact for the most part with undamaged DNA. Whereas the structural basis for recognition of DNA lesions by glycosylases has been studied extensively, less is known about the nature of the interaction between these proteins and undamaged DNA. Here we report the crystal structures of the DNA glycosylase AlkA in complex with undamaged DNA. The structures revealed a recognition mode in which the DNA is nearly straight, with no amino acid side chains inserted into the duplex, and the target base pair is fully intrahelical. A comparison of the present structures with that of AlkA recognizing an extrahelical lesion revealed conformational changes in both the DNA and protein as the glycosylase transitions from the interrogation of undamaged DNA to catalysis of nucleobase excision. Modeling studies with the cytotoxic lesion 3-methyladenine and accompanying biochemical experiments suggested that AlkA actively interrogates the minor groove of the DNA while probing for the presence of lesions.
- Department of Stem Cell and Regenerative Biology, Harvard University, Cambridge, Massachusetts 02138, USA.
Organizational Affiliation: