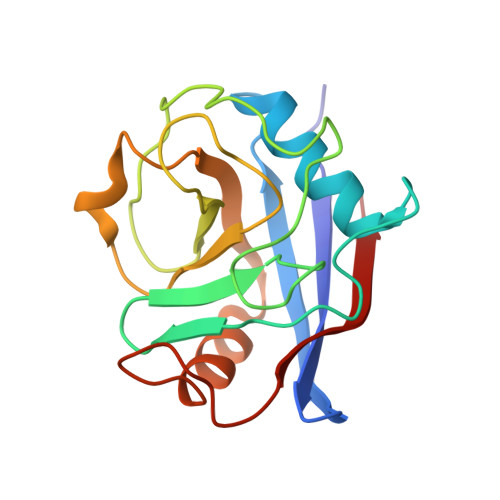
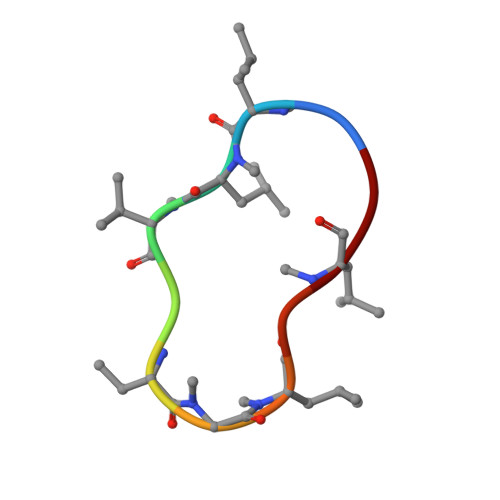
Structural basis for the cyclophilin A binding affinity and immunosuppressive potency of E-ISA247 (voclosporin).
Kuglstatter, A., Mueller, F., Kusznir, E., Gsell, B., Stihle, M., Thoma, R., Benz, J., Aspeslet, L., Freitag, D., Hennig, M.(2011) Acta Crystallogr D Biol Crystallogr 67: 119-123
- PubMed: 21245533
- DOI: https://doi.org/10.1107/S0907444910051905
- Primary Citation of Related Structures:
3ODI, 3ODL - PubMed Abstract:
E-ISA247 (voclosporin) is a cyclosporin A analogue that is in late-stage clinical development for the treatment of autoimmune diseases and the prevention of organ graft rejection. The X-ray crystal structures of E-ISA247 and its stereoisomer Z-ISA247 bound to cyclophilin A have been determined and their binding affinities were measured to be 15 and 61 nM, respectively, by fluorescence spectroscopy. The higher affinity of E-ISA247 can be explained by superior van der Waals contacts between its unique side chain and cyclophilin A. Comparison with the known ternary structure including calcineurin suggests that the higher immunosuppressive efficacy of E-ISA247 relative to cyclosporin A could be a consequence of structural changes in calcineurin induced by the modified E-ISA247 side chain.
- F. Hoffmann-La Roche, Discovery Technologies, 4070 Basel, Switzerland.
Organizational Affiliation: