Fab'-induced folding of antigenic N-terminal peptides from intrinsically disordered HIV-1 Tat revealed by X-ray crystallography.
Serriere, J., Dugua, J.M., Bossus, M., Verrier, B., Haser, R., Gouet, P., Guillon, C.(2011) J Mol Biology 405: 33-42
- PubMed: 21035463
- DOI: https://doi.org/10.1016/j.jmb.2010.10.033
- Primary Citation of Related Structures:
3O6K, 3O6L, 3O6M - PubMed Abstract:
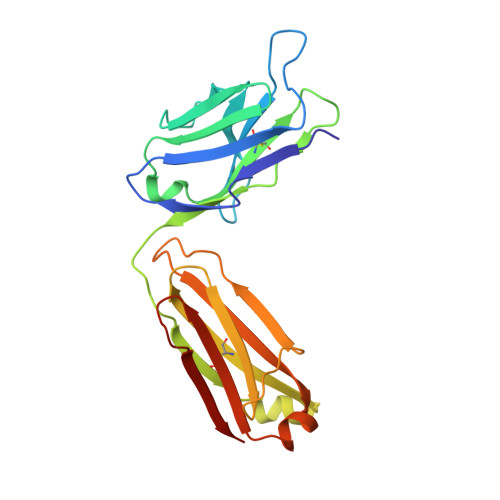
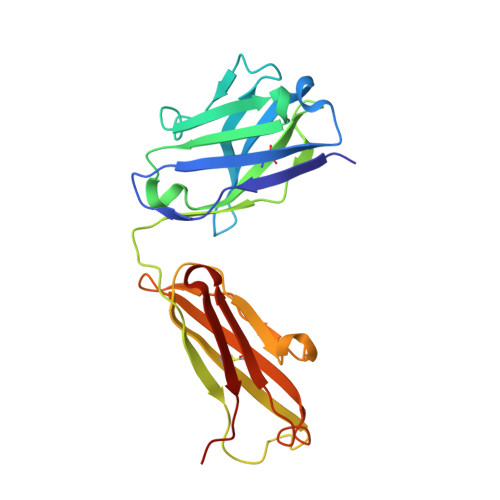
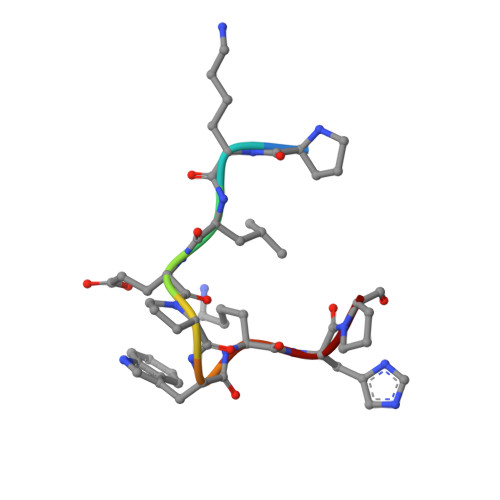
Tat, the transcriptional activator protein of human immunodeficiency virus type 1 (HIV-1), is critical for viral replication and is a potential HIV-1 vaccine candidate. This intrinsically disordered protein is present in the extracellular medium and is involved in the pathogenicity of HIV through its interaction with different cellular and viral biological partners. A monoclonal antibody termed 11H6H1, which is specific for the N-terminal region of Tat, was selected for a functional and structural study of the HIV-1 Tat protein. The equilibrium dissociation constants (K(d)) of Tat and Tat fragments complexed with 11H6H1 were estimated by competitive ELISA. Tat contains a single tryptophan residue, Trp11, located in the N-terminal region. We show that the substitution of Trp11 by a phenylalanine completely abolishes the binding of 11H6H1, whereas the transactivating activity of Tat is preserved. The epitope recognized by 11H6H1 was restricted to the 9-mer peptide P(6)KLEPWKHP(14) centered on Trp11. The crystal structures of this 9-mer peptide and of an overlapping 15-mer peptide were determined in complex with Fab' 11H6H1 at 2.4 Å and 2.1 Å resolution, respectively. Tat is intrinsically disordered and can undergo induced folding upon association with a biological partner. Our crystallographic study reveals that the two Tat peptides, which are lodged in the U-shaped groove of the Fab' antigen-binding site, adopt a standard type I β-turn conformation. The central Trp11 that is critical for Fab' recognition is further stabilized by π-stacking interactions. The structural and biological consequences of this induced folding in HIV pathogenesis are discussed.
- Université de Lyon, IFR128 BioSciences Gerland-Lyon Sud, 7 Passage du Vercors, France.
Organizational Affiliation: