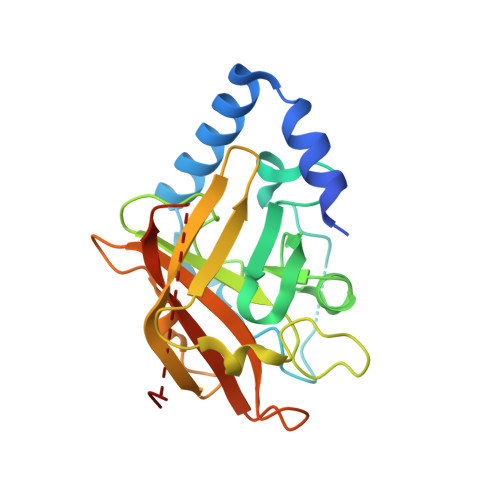
Structure analysis and site-directed mutagenesis of defined key residues and motives for pilus-related sortase C1 in group B Streptococcus.
Cozzi, R., Malito, E., Nuccitelli, A., D'Onofrio, M., Martinelli, M., Ferlenghi, I., Grandi, G., Telford, J.L., Maione, D., Rinaudo, C.D.(2011) FASEB J 25: 1874-1886
- PubMed: 21357525
- DOI: https://doi.org/10.1096/fj.10-174797
- Primary Citation of Related Structures:
3O0P - PubMed Abstract:
In group B Streptococcus (GBS), 3 structurally distinct types of pili have been discovered as potential virulence factors and vaccine candidates. The pilus-forming proteins are assembled into high-molecular-weight polymers via a transpeptidation mechanism mediated by specific class C sortases. Using a multidisciplinary approach including bioinformatics, structural and biochemical studies, and in vivo mutagenesis, we performed a broad characterization of GBS sortase C1 of pilus island 2a. The high-resolution X-ray structure of the enzyme revealed that the active site, into the β-barrel core of the enzyme, is made of the catalytic triad His157-Cys219-Arg228 and covered by a loop, known as the "lid." We show that the catalytic triad and the predicted N- and C-terminal transmembrane regions are required for the enzyme activity. Interestingly, by in vivo complementation mutagenesis studies, we found that the deletion of the entire lid loop or mutations in specific lid key residues had no effect on catalytic activity of the enzyme. In addition, kinetic characterizations of recombinant enzymes indicate that the lid mutants can still recognize and cleave the substrate-mimicking peptide at least as well as the wild-type protein.
- Novartis Vaccines and Diagnostics, Via Fiorentina 1, 53100 Siena, Italy.
Organizational Affiliation: