Metronidazole Activation by a Deeply Entangled Dimeric Malic Enzyme in Entamoeba histolytica.
Chakrabarty, A., Dutta, D., Baidya, M., Dutta, A., Das, A.K., Ghosh, S.K.(2025) Pathogens 14
- PubMed: 40137762
- DOI: https://doi.org/10.3390/pathogens14030277
- Primary Citation of Related Structures:
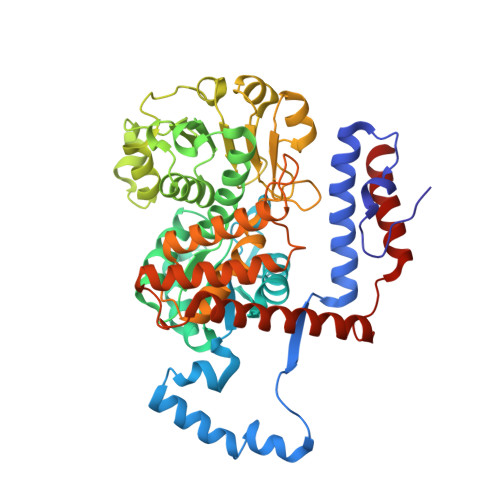
3NV9 - PubMed Abstract:
Metronidazole is the preferred drug for treating amoebiasis caused by Entamoeba histolytica . Its antiamoebic activity is primarily attributed to activation by various reductases. This study reports an alternative activation pathway in E. histolytica mediated by the decarboxylating malic enzyme. Functional characterization of this NADPH-dependent enzyme reveals that it is secreted into the extracellular milieu and may play a role in E. histolytica adhesion to human enteric cells. Structural analysis of the E. histolytica malic enzyme (EhME) demonstrates that the protein forms a strict dimer, with the protomers interlocked by a unique knot structure formed by two polypeptide chains. This distinctive structural feature closely aligns EhME with its prokaryotic counterparts. In conclusion, our findings reveal that E. histolytica harbors a deeply entangled dimeric malic enzyme that contributes to metronidazole susceptibility, sharing structural similarities with bacterial malic enzymes.
- Department of Bioscience and Biotechnology, Indian Institute of Technology Kharagpur, Kharagpur 721302, India.
Organizational Affiliation: