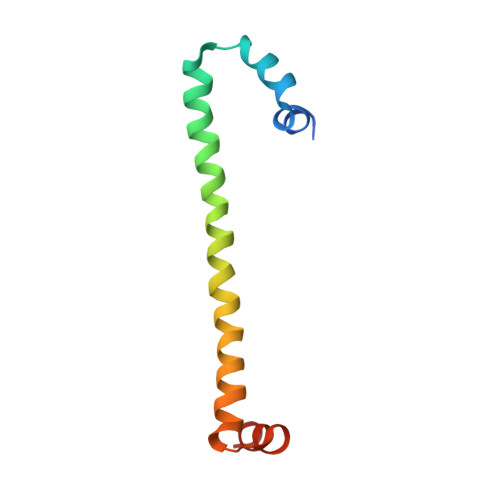
H-NS forms a superhelical protein scaffold for DNA condensation.
Arold, S.T., Leonard, P.G., Parkinson, G.N., Ladbury, J.E.(2010) Proc Natl Acad Sci U S A 107: 15728-15732
- PubMed: 20798056
- DOI: https://doi.org/10.1073/pnas.1006966107
- Primary Citation of Related Structures:
3NR7 - PubMed Abstract:
The histone-like nucleoid structuring (H-NS) protein plays a fundamental role in DNA condensation and is a key regulator of enterobacterial gene expression in response to changes in osmolarity, pH, and temperature. The protein is capable of high-order self-association via interactions of its oligomerization domain. Using crystallography, we have solved the structure of this complete domain in an oligomerized state. The observed superhelical structure establishes a mechanism for the self-association of H-NS via both an N-terminal antiparallel coiled-coil and a second, hitherto unidentified, helix-turn-helix dimerization interface at the C-terminal end of the oligomerization domain. The helical scaffold suggests the formation of a H-NS:plectonemic DNA nucleoprotein complex that is capable of explaining published biophysical and functional data, and establishes a unifying structural basis for coordinating the DNA packaging and transcription repression functions of H-NS.
- Department of Biochemistry and Molecular Biology, University of Texas, M. D. Anderson Cancer Center, Houston, 77030, USA.
Organizational Affiliation: