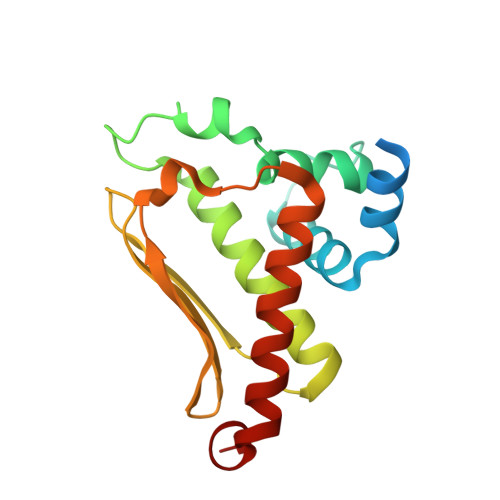
Crystal structure of cce_0566 from Cyanothece 51142, a protein associated with nitrogen fixation in the DUF269 family.
Buchko, G.W., Robinson, H.(2012) FEBS Lett 586: 350-355
- PubMed: 22289180
- DOI: https://doi.org/10.1016/j.febslet.2012.01.037
- Primary Citation of Related Structures:
3NJ2 - PubMed Abstract:
The crystal structure for cce_0566 (171 aa, 19.4 kDa), a DUF269 annotated protein from the diazotrophic cyanobacterium Cyanothece sp. ATCC 51142, was determined to 1.60Å resolution. Cce_0566 is a homodimer with each molecule composed of eight α-helices folded on one side of a three strand anti-parallel β-sheet. Hydrophobic interactions between the side chains of largely conserved residues on the surface of each β-sheet hold the dimer together. The fold observed for cce_0566 may be unique to proteins in the DUF269 family, hence, the protein may also have a function unique to nitrogen fixation. A solvent accessible cleft containing conserved charged residues near the dimer interface could represent the active site or ligand-binding surface for the protein's biological function.
- Biological Sciences Division, Pacific Northwest National Laboratory, Richland, WA 99352, USA. garry.buchko@pnnl.gov
Organizational Affiliation: