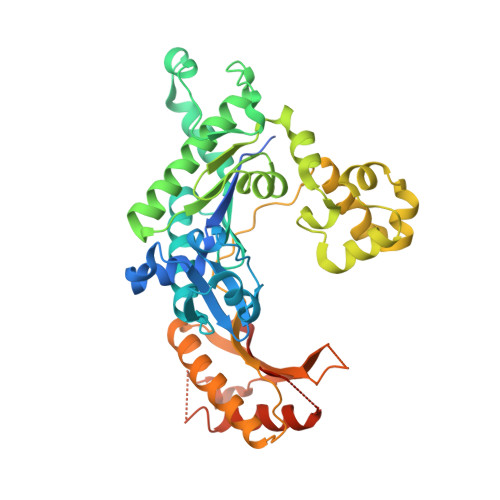
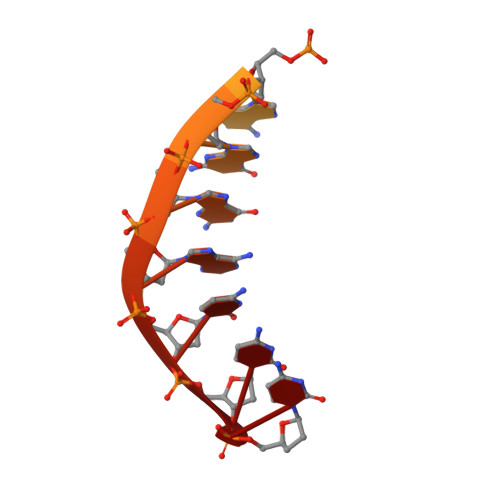
Structural Basis for Proficient Incorporation of dTTP Opposite O6-Methylguanine by Human DNA Polymerase {iota}.
Pence, M.G., Choi, J.Y., Egli, M., Guengerich, F.P.(2010) J Biological Chem 285: 40666-40672
- PubMed: 20961860
- DOI: https://doi.org/10.1074/jbc.M110.183665
- Primary Citation of Related Structures:
3NGD, 3OSN - PubMed Abstract:
O(6)-methylguanine (O(6)-methylG) is highly mutagenic and is commonly found in DNA exposed to methylating agents, even physiological ones (e.g. S-adenosylmethionine). The efficiency of a truncated, catalytic DNA polymerase ι core enzyme was determined for nucleoside triphosphate incorporation opposite O(6)-methylG, using steady-state kinetic analyses. The results presented here corroborate previous work from this laboratory using full-length pol ι, which showed that dTTP incorporation occurs with high efficiency opposite O(6)-methylG. Misincorporation of dTTP opposite O(6)-methylG occurred with ∼6-fold higher efficiency than incorporation of dCTP. Crystal structures of the truncated form of pol ι with O(6)-methylG as the template base and incoming dCTP or dTTP were solved and showed that O(6)-methylG is rotated into the syn conformation in the pol ι active site and that dTTP misincorporation by pol ι is the result of Hoogsteen base pairing with the adduct. Both dCTP and dTTP base paired with the Hoogsteen edge of O(6)-methylG. A single, short hydrogen bond formed between the N3 atom of dTTP and the N7 atom of O(6)-methylG. Protonation of the N3 atom of dCTP and bifurcation of the N3 hydrogen between the N7 and O(6) atoms of O(6)-methylG allow base pairing of the lesion with dCTP. We conclude that differences in the Hoogsteen hydrogen bonding between nucleotides is the main factor in the preferential selectivity of dTTP opposite O(6)-methylG by human pol ι, in contrast to the mispairing modes observed previously for O(6)-methylG in the structures of the model DNA polymerases Sulfolobus solfataricus Dpo4 and Bacillus stearothermophilus DNA polymerase I.
- Department of Biochemistry and Center in Molecular Toxicology, Vanderbilt University School of Medicine, Nashville, Tennessee 37232-0146, USA.
Organizational Affiliation: