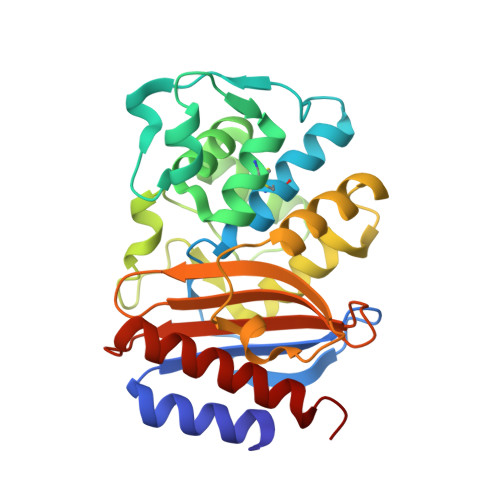
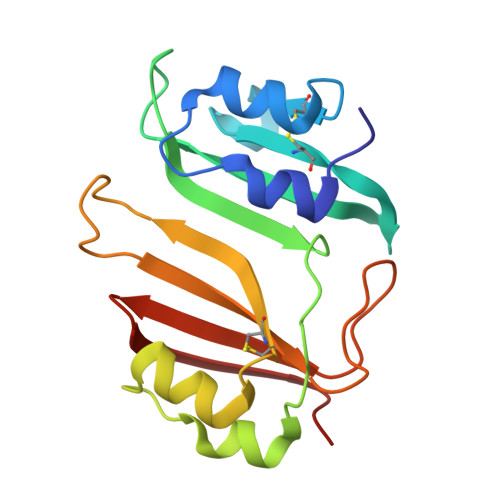
Specificity and Cooperativity at beta-lactamase Position 104 in TEM-1/BLIP and SHV-1/BLIP interactions
Hanes, M.S., Reynolds, K.A., McNamara, C., Ghosh, P., Bonomo, R.A., Kirsch, J.F., Handel, T.M.(2011) Proteins 79: 1267-1276
- PubMed: 21294157
- DOI: https://doi.org/10.1002/prot.22961
- Primary Citation of Related Structures:
3N4I - PubMed Abstract:
Establishing a quantitative understanding of the determinants of affinity in protein-protein interactions remains challenging. For example, TEM-1/β-lactamase inhibitor protein (BLIP) and SHV-1/BLIP are homologous β-lactamase/β-lactamase inhibitor protein complexes with disparate K(d) values (3 nM and 2 μM, respectively), and a single substitution, D104E in SHV-1, results in a 1000-fold enhancement in binding affinity. In TEM-1, E104 participates in a salt bridge with BLIP K74, whereas the corresponding SHV-1 D104 does not in the wild type SHV-1/BLIP co-structure. Here, we present a 1.6 Å crystal structure of the SHV-1 D104E/BLIP complex that demonstrates that this point mutation restores this salt bridge. Additionally, mutation of a neighboring residue, BLIP E73M, results in salt bridge formation between SHV-1 D104 and BLIP K74 and a 400-fold increase in binding affinity. To understand how this salt bridge contributes to complex affinity, the cooperativity between the E/K or D/K salt bridge pair and a neighboring hot spot residue (BLIP F142) was investigated using double mutant cycle analyses in the background of the E73M mutation. We find that BLIP F142 cooperatively stabilizes both interactions, illustrating how a single mutation at a hot spot position can drive large perturbations in interface stability and specificity through a cooperative interaction network.
- Biophysics Graduate Group, University of California, Berkeley, Berkeley, California 94729, USA.
Organizational Affiliation: