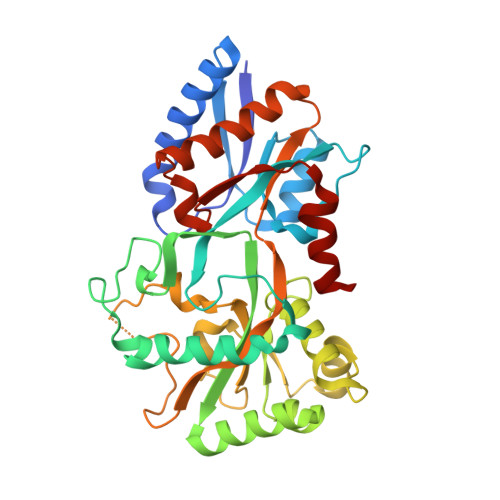
Insights into Mycoplasma genitalium metabolism revealed by the structure of MG289, an extracytoplasmic thiamine binding lipoprotein.
Sippel, K.H., Venkatakrishnan, B., Boehlein, S.K., Sankaran, B., Quirit, J.G., Govindasamy, L., Agbandje-McKenna, M., Goodison, S., Rosser, C.J., McKenna, R.(2011) Proteins 79: 528-536
- PubMed: 21117240
- DOI: https://doi.org/10.1002/prot.22900
- Primary Citation of Related Structures:
3MYU - PubMed Abstract:
Mycoplasma genitalium is one of the smallest organisms capable of self-replication and its sequence is considered a starting point for understanding the minimal genome required for life. MG289, a putative phosphonate substrate binding protein, is considered to be one of these essential genes. The crystal structure of MG289 has been solved at 1.95 Å resolution. The structurally identified thiamine binding region reveals possible mechanisms for ligand promiscuity. MG289 was determined to be an extracytoplasmic thiamine binding lipoprotein. Computational analysis, size exclusion chromatography, and small angle X-ray scattering indicates that MG289 homodimerizes in a concentration-dependant manner. Comparisons to the thiamine pyrophosphate binding homolog Cypl reveal insights into the metabolic differences between mycoplasmal species including identifying possible kinases for cofactor phosphorylation and describing the mechanism of thiamine transport into the cell. These results provide a baseline to build our understanding of the minimal metabolic requirements of a living organism.
- Department of Biochemistry and Molecular Biology, College of Medicine, Gainesville, Florida 32610, USA.
Organizational Affiliation: