An allosteric inhibitor of substrate recognition by the SCF(Cdc4) ubiquitin ligase.
Orlicky, S., Tang, X., Neduva, V., Elowe, N., Brown, E.D., Sicheri, F., Tyers, M.(2010) Nat Biotechnol 28: 733-737
- PubMed: 20581844
- DOI: https://doi.org/10.1038/nbt.1646
- Primary Citation of Related Structures:
3MKS - PubMed Abstract:
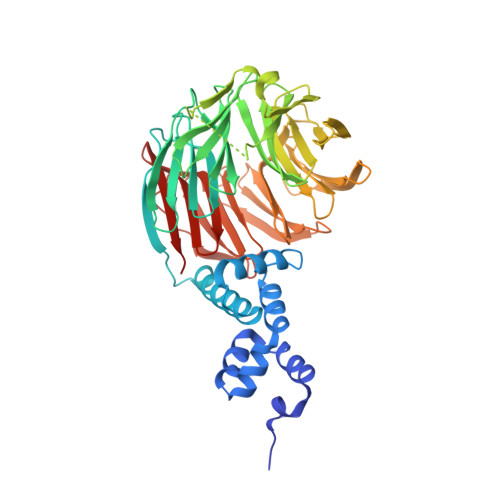
The specificity of SCF ubiquitin ligase-mediated protein degradation is determined by F-box proteins. We identified a biplanar dicarboxylic acid compound, called SCF-I2, as an inhibitor of substrate recognition by the yeast F-box protein Cdc4 using a fluorescence polarization screen to monitor the displacement of a fluorescein-labeled phosphodegron peptide. SCF-I2 inhibits the binding and ubiquitination of full-length phosphorylated substrates by SCF(Cdc4). A co-crystal structure reveals that SCF-I2 inserts itself between the beta-strands of blades 5 and 6 of the WD40 propeller domain of Cdc4 at a site that is 25 A away from the substrate binding site. Long-range transmission of SCF-I2 interactions distorts the substrate binding pocket and impedes recognition of key determinants in the Cdc4 phosphodegron. Mutation of the SCF-I2 binding site abrogates its inhibitory effect and explains specificity in the allosteric inhibition mechanism. Mammalian WD40 domain proteins may exhibit similar allosteric responsiveness and hence represent an extensive class of druggable target.
- Center for Systems Biology, Samuel Lunenfeld Research Institute, Mount Sinai Hospital, Toronto, Canada.
Organizational Affiliation: