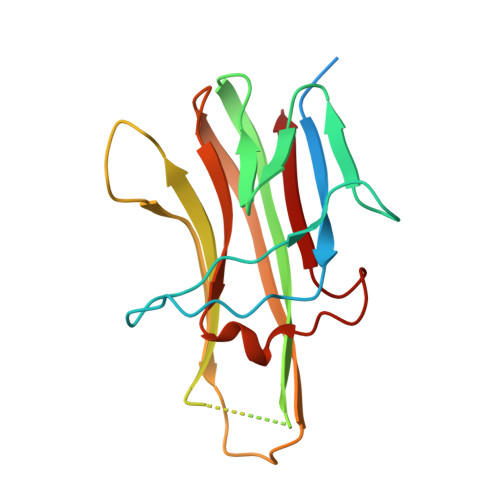
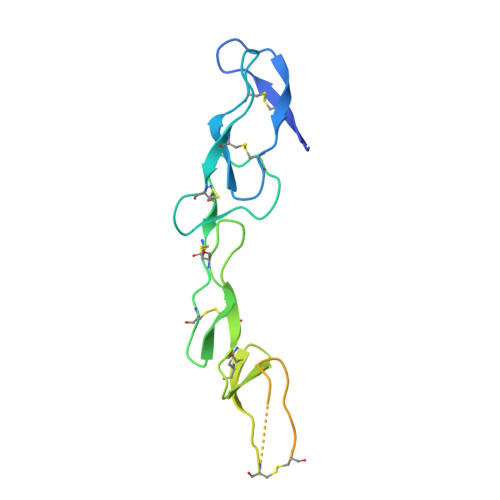
Decoy Strategies: The Structure of TL1A:DcR3 Complex.
Zhan, C., Patskovsky, Y., Yan, Q., Li, Z., Ramagopal, U., Cheng, H., Brenowitz, M., Hui, X., Nathenson, S.G., Almo, S.C.(2011) Structure 19: 162-171
- PubMed: 21300286
- DOI: https://doi.org/10.1016/j.str.2010.12.004
- Primary Citation of Related Structures:
3K51, 3MHD, 3MI8 - PubMed Abstract:
Decoy Receptor 3 (DcR3), a secreted member of the Tumor Necrosis Factor (TNF) receptor superfamily, neutralizes three different TNF ligands: FasL, LIGHT, and TL1A. Each of these ligands engages unique signaling receptors which direct distinct and critical immune responses. We report the crystal structures of the unliganded DcR3 ectodomain and its complex with TL1A, as well as complementary mutagenesis and biochemical studies. These analyses demonstrate that DcR3 interacts with invariant backbone and side-chain atoms in the membrane-proximal half of TL1A which supports recognition of its three distinct TNF ligands. Additional features serve as antideterminants that preclude interaction with other members of the TNF superfamily. This mode of interaction is unique among characterized TNF:TNFR family members and provides a mechanistic basis for the broadened specificity required to support the decoy function of DcR3, as well as for the rational manipulation of specificity and affinity of DcR3 and its ligands.
- Department of Biochemistry, Albert Einstein College of Medicine, Bronx, NY 10461, USA.
Organizational Affiliation: