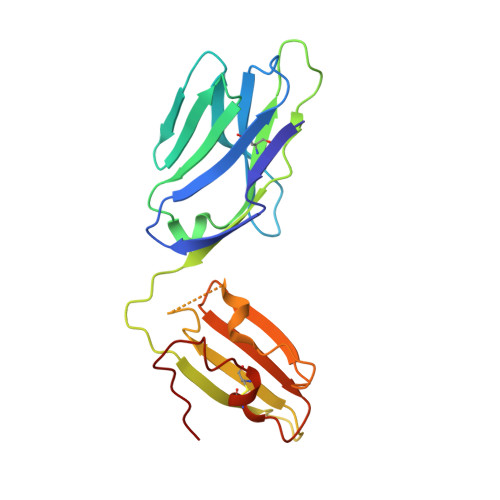
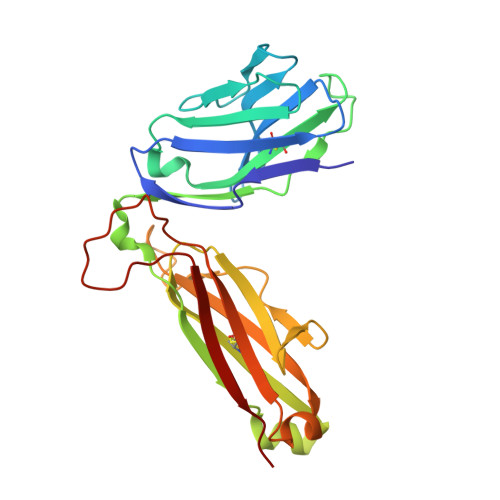
An alternative conformation of the T-cell receptor alpha constant region
van Boxel, G.I., Holmes, S., Fugger, L., Jones, E.Y.(2010) J Mol Biology 400: 828-837
- PubMed: 20630474
- DOI: https://doi.org/10.1016/j.jmb.2010.05.053
- Primary Citation of Related Structures:
3MFF - PubMed Abstract:
Alphabeta T-cell receptors (TcRs) play a central role in cellular immune response. They are members of the Ig superfamily, with extracellular regions of the alpha and beta chains each comprising a V-type domain and a C-type domain. We have determined the ectodomain structure of an alphabeta TcR, which recognizes the autoantigen myelin basic protein. The 2.0-A-resolution structure reveals canonical main-chain conformations for the V(alpha), V(beta), and C(beta) domains, but the C(alpha) domain exhibits a main-chain conformation remarkably different from those previously reported for TcR crystal structures. The global IgC-like fold is maintained, but a piston-like rearrangement between BC and DE beta-turns results in beta-strand slippage. This substantial conformational change may represent a signaling intermediate. Our structure is the first example for the Ig fold of the increasingly recognized concept of "metamorphic proteins."
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, The University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Organizational Affiliation: