Crystal Structure of Geranylgeranyl Pyrophosphate Synthase from Lactobacillus Brevis Atcc 367
Patskovsky, Y., Toro, R., Rutter, M., Sauder, J.M., Burley, S.K., Almo, S.C.To be published.
Experimental Data Snapshot
wwPDB Validation 3D Report Full Report
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Farnesyl-diphosphate synthase | 309 | Levilactobacillus brevis ATCC 367 | Mutation(s): 0 Gene Names: LVIS_0975 EC: 2.5.1.10 | 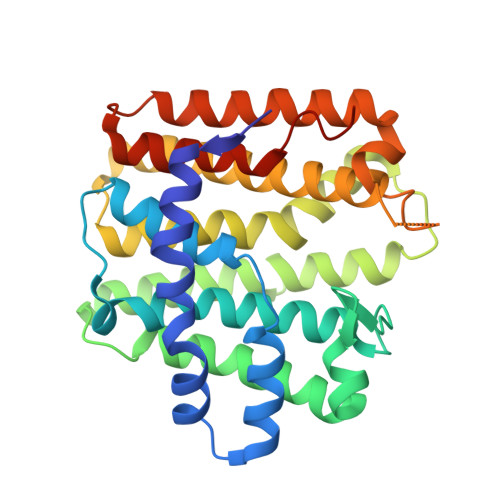 | |
UniProt | |||||
Find proteins for Q03RR4 (Levilactobacillus brevis (strain ATCC 367 / BCRC 12310 / CIP 105137 / JCM 1170 / LMG 11437 / NCIMB 947 / NCTC 947)) Explore Q03RR4 Go to UniProtKB: Q03RR4 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q03RR4 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 1 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| GOL Query on GOL | E [auth A] F [auth A] G [auth A] H [auth A] I [auth A] | GLYCEROL C3 H8 O3 PEDCQBHIVMGVHV-UHFFFAOYSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 48.496 | α = 95.47 |
| b = 51.096 | β = 91.28 |
| c = 125.928 | γ = 105.56 |
| Software Name | Purpose |
|---|---|
| SHELX | model building |
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| SHELX | phasing |