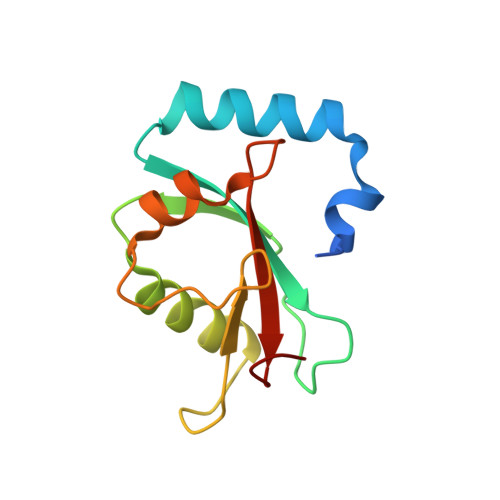
Structure of autophagy-related protein Atg8 from the silkworm Bombyx mori
Hu, C., Zhang, X., Teng, Y.-B., Hu, H.-X., Li, W.-F.(2010) Acta Crystallogr Sect F Struct Biol Cryst Commun 66: 787-790
- PubMed: 20606273
- DOI: https://doi.org/10.1107/S1744309110018464
- Primary Citation of Related Structures:
3M95 - PubMed Abstract:
Autophagy-related protein Atg8 is ubiquitous in all eukaryotes. It is involved in the Atg8-PE ubiquitin-like conjugation system, which is essential for autophagosome formation. The structures of Atg8 from different species are very similar and share a ubiquitin-fold domain at the C-terminus. In the 2.40 A crystal structure of Atg8 from the silkworm Bombyx mori reported here, the ubiquitin fold at the C-terminus is preceded by two additional helices at the N-terminus.
- Hefei National Laboratory for Physical Sciences at Microscale and School of Life Sciences, University of Science and Technology of China, People's Republic of China.
Organizational Affiliation: